3R94
 
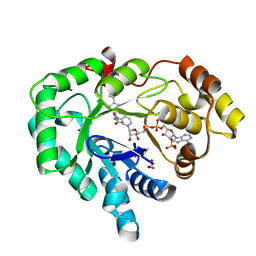 | | AKR1C3 complex with flurbiprofen | | Descriptor: | (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, ... | | Authors: | Yosaatmadja, Y, Teague, R.M, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2011-03-24 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.013 Å) | | Cite: | Crystal structures of three classes of non-steroidal anti-inflammatory drugs in complex with aldo-keto reductase 1C3.
Plos One, 7, 2012
|
|
3CZS
 
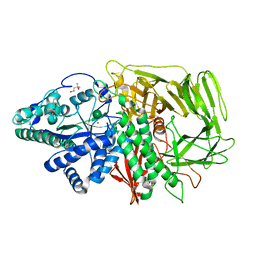 | | Golgi alpha-mannosidase II (D204A nucleophile mutant) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-mannosidase 2, ... | | Authors: | Shah, N, Rose, D.R. | | Deposit date: | 2008-04-29 | | Release date: | 2008-06-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Golgi alpha-mannosidase II cleaves two sugars sequentially in the same catalytic site.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1QJA
 
 | | 14-3-3 ZETA/PHOSPHOPEPTIDE COMPLEX (MODE 2) | | Descriptor: | 14-3-3 PROTEIN ZETA, PHOSPHOPEPTIDE | | Authors: | Rittinger, K, Budman, J, Xu, J, Volinia, S, Cantley, L.C, Smerdon, S.J, Gamblin, S.J, Yaffe, M.B. | | Deposit date: | 1999-06-23 | | Release date: | 1999-09-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of 14-3-3 Phosphopeptide Complexes Identifies a Dual Role for the Nuclear Export Signal of 14-3-3 in Ligand Binding
Mol.Cell, 4, 1999
|
|
3TE7
 
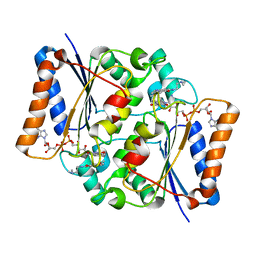 | | Quinone Oxidoreductase (NQ02) bound to the imidazoacridin-6-one 5a1 | | Descriptor: | 5-{[2-(dimethylamino)ethyl]amino}-8-methoxy-6H-imidazo[4,5,1-de]acridin-6-one, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Dunstan, M.S. | | Deposit date: | 2011-08-12 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel Inhibitors of NRH:Quinone Oxidoreductase 2 (NQO2): Crystal Structures, Biochemical Activity, and Intracellular Effects of Imidazoacridin-6-ones.
J.Med.Chem., 54, 2011
|
|
3D60
 
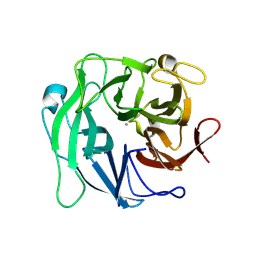 | | Crystal Structure Analysis of 1,5-alpha-arabinanase catalytic mutant (D27A) | | Descriptor: | CALCIUM ION, Intracellular arabinanase | | Authors: | Alhassid, A, Ben David, A, Shoham, Y, Shoham, G. | | Deposit date: | 2008-05-18 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an inverting GH 43 1,5-alpha-L-arabinanase from Geobacillus stearothermophilus complexed with its substrate
Biochem.J., 422, 2009
|
|
1PM9
 
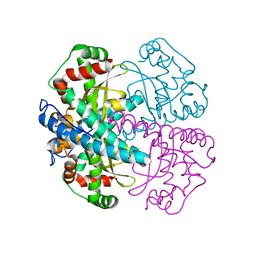 | | CRYSTAL STRUCTURE OF HUMAN MNSOD H30N, Y166F MUTANT | | Descriptor: | MANGANESE (III) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Fan, L, Tainer, J.A. | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Amino acid substitution at the dimeric interface of human manganese superoxide dismutase
J.Biol.Chem., 279, 2004
|
|
3AMD
 
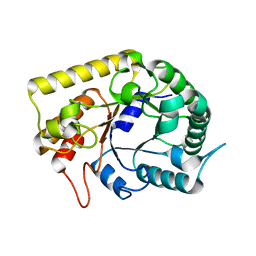 | | Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3THJ
 
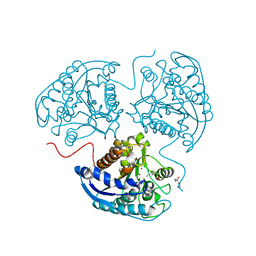 | |
3A0H
 
 | | Crystal structure of I-substituted Photosystem II complex | | Descriptor: | (1S)-2-(ALPHA-L-ALLOPYRANOSYLOXY)-1-[(TRIDECANOYLOXY)METHYL]ETHYL PALMITATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 5-[(2E,6E,10E,14E,18E,22E)-3,7,11,15,19,23,27-HEPTAMETHYLOCTACOSA-2,6,10,14,18,22,26-HEPTAENYL]-2,3-DIMETHYLBENZO-1,4-QUINONE, ... | | Authors: | Kawakami, K, Umena, Y, Kamiya, N, Shen, J.-R. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Location of chloride and its possible functions in oxygen-evolving photosystem II revealed by X-ray crystallography
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3TDK
 
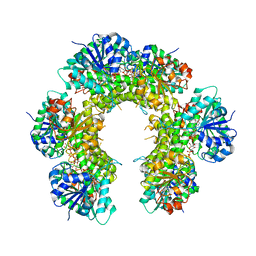 | |
3TH1
 
 | | Crystal structure of chlorocatechol 1,2-dioxygenase from Pseudomonas putida | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ACETATE ION, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Rustiguel, J.K, Nonato, M.C. | | Deposit date: | 2011-08-18 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of chlorocatechol 1,2-dioxygenase from Pseudomonas putida
To be Published
|
|
3THX
 
 | | Human MutSbeta complexed with an IDL of 3 bases (Loop3) and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA Loop3 minus strand, DNA Loop3 plus strand, ... | | Authors: | Yang, W. | | Deposit date: | 2011-08-19 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of mismatch recognition revealed by human MutSbeta bound to unpaired DNA loops
Nat.Struct.Mol.Biol., 19, 2012
|
|
3A6E
 
 | | W174F mutant creatininase, type I | | Descriptor: | CACODYLATE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A4J
 
 | | arPTE (K185R/D208G/N265D/T274N) | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Phosphotriesterase | | Authors: | Foo, J.L, Jackson, C.J, Carr, P.D, Ollis, D.L. | | Deposit date: | 2009-07-07 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Conformational sampling, catalysis, and evolution of the bacterial phosphotriesterase
Proc.Natl.Acad.Sci.USA, 2009
|
|
3A96
 
 | | Crystal structure of hen egg white lysozyme soaked with 100mM RhCl3 at pH2.2 | | Descriptor: | CHLORIDE ION, Lysozyme C, RHODIUM(III) ION, ... | | Authors: | Abe, S, Koshiyama, T, Ohki, T, Hikage, T, Watanabe, Y, Ueno, T. | | Deposit date: | 2009-10-15 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elucidation of Metal-Ion Accumulation Induced by Hydrogen Bonds on Protein Surfaces by Using Porous Lysozyme Crystals Containing Rh(III) Ions as the Model Surfaces
Chemistry, 16, 2010
|
|
3A69
 
 | |
1PML
 
 | |
3A6K
 
 | | The E122Q mutant creatininase, Mn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3AEF
 
 | | Crystal structure of porcine heart mitochondrial complex II with an empty quinone-binding pocket | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Harada, S, Sasaki, T, Shindo, M, Kido, Y, Inaoka, D.K, Omori, J, Osanai, A, Sakamoto, K, Mao, J, Matsuoka, S, Inoue, M, Honma, T, Tanaka, A, Kita, K. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of porcine heart mitochondrial complex II with an empty quinone-binding pocket
To be Published
|
|
3A8R
 
 | | The structure of the N-terminal regulatory domain of a plant NADPH oxidase | | Descriptor: | CALCIUM ION, Putative uncharacterized protein | | Authors: | Oda, T, Hashimoto, H, Kuwabara, N, Akashi, S, Hayashi, K, Kojima, C, Wong, H.L, Kawasaki, T, Shimamoto, K, Sato, M, Shimizu, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-10-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications
J.Biol.Chem., 285, 2010
|
|
3THY
 
 | | Human MutSbeta complexed with an IDL of 2 bases (Loop2) and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA Loop2 minus strand, DNA Loop2 plus strand, ... | | Authors: | Yang, W. | | Deposit date: | 2011-08-19 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Mechanism of mismatch repair revealed by human MutSbeta bound to unpaired DNA loops
Nat.Struct.Mol.Biol., 19, 2012
|
|
3A9B
 
 | | CcCel6C, a glycoside hydrolase family 6 enzyme, complexed with cellobiose | | Descriptor: | Cellobiohydrolase, MAGNESIUM ION, beta-D-glucopyranose, ... | | Authors: | Liu, Y, Yoshida, M, Kurakata, Y, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | Deposit date: | 2009-10-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of a glycoside hydrolase family 6 enzyme, CcCel6C, a cellulase constitutively produced by Coprinopsis cinerea
Febs J., 277, 2010
|
|
3TKK
 
 | | Crystal Structure Analysis of a recombinant predicted acetamidase/ formamidase from the thermophile thermoanaerobacter tengcongensis | | Descriptor: | CALCIUM ION, Predicted acetamidase/formamidase, ZINC ION | | Authors: | Qian, M, Huang, Q, Wu, G, Lai, L, Tang, Y, Pei, J, Kusunoki, M. | | Deposit date: | 2011-08-26 | | Release date: | 2011-11-16 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure Analysis of a Recombinant Predicted Acetamidase/Formamidase from the Thermophile Thermoanaerobacter tengcongensis.
PROTEIN J., 31, 2012
|
|
3AIW
 
 | | Crystal structure of beta-glucosidase in rye complexed with 2-deoxy-2-fluoroglucoside and dinitrophenol | | Descriptor: | 2,4-DINITROPHENOL, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
3AJ7
 
 | | Crystal Structure of isomaltase from Saccharomyces cerevisiae | | Descriptor: | CALCIUM ION, Oligo-1,6-glucosidase | | Authors: | Yamamoto, K, Miyake, H, Kusunoki, M, Osaki, S. | | Deposit date: | 2010-05-26 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of isomaltase from Saccharomyces cerevisiae and in complex with its competitive inhibitor maltose
Febs J., 277, 2010
|
|
