1QNT
 
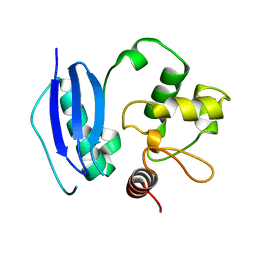 | |
222D
 
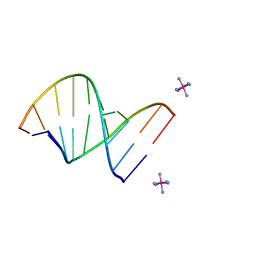 | | INFLUENCE OF COUNTER-IONS ON THE CRYSTAL STRUCTURES OF DNA DECAMERS: BINDING OF [CO(NH3)6]3+ AND BA2+ TO A-DNA | | Descriptor: | COBALT HEXAMMINE(III), DNA/RNA (5'-R(*GP*CP*)-D(*GP*TP*AP*TP*AP*CP*GP*C)-3') | | Authors: | Gao, Y.-G, Robinson, H, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1995-06-26 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA.
Biophys.J., 69, 1995
|
|
1A9I
 
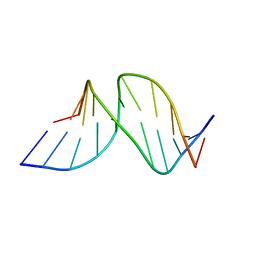 | |
1A9J
 
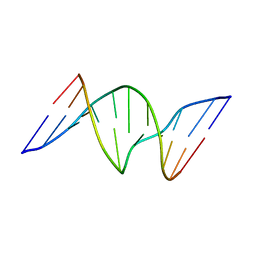 | |
206D
 
 | |
254D
 
 | |
1TDZ
 
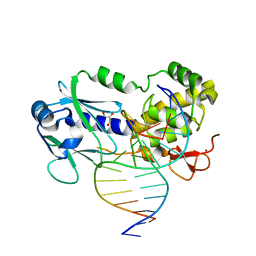 | | Crystal Structure Complex Between the Lactococcus Lactis FPG (Mutm) and a FAPY-dG Containing DNA | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(FOX)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', GLYCEROL, ... | | Authors: | Coste, F, Ober, M, Carell, T, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2004-05-24 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
275D
 
 | |
2HLI
 
 | | Solution structure of Crotonaldehyde-Derived N2-[3-Oxo-1(S)-methyl-propyl]-dG DNA Adduct in the 5'-CpG-3' Sequence | | Descriptor: | DNA dodecamer, DNA dodecamer with S-crotonaldehyde adduct | | Authors: | Cho, Y.-J, Wang, H, Kozekov, I.D, Kurtz, A.J, Jacob, J, Voehler, M, Smith, J, Harris, T.M, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Orientation of the Crotonaldehyde-Derived N(2)-[3-Oxo-1(S)-methyl-propyl]-dG DNA Adduct Hinders Interstrand Cross-Link Formation in the 5'-CpG-3' Sequence
Chem.Res.Toxicol., 19, 2006
|
|
3QK4
 
 | |
1BUA
 
 | |
260D
 
 | |
8S9M
 
 | | DNA cytosine-N4 methyltransferase (residues 79-324) from the Bdelloid rotifer Adineta vaga | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9N
 
 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - C2 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
8S9O
 
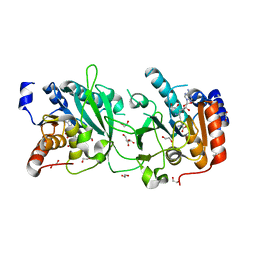 | | DNA cytosine-N4 methyltransferase (residues 61-324) from the Bdelloid rotifer Adineta vaga - P1 crystal form | | Descriptor: | 1,2-ETHANEDIOL, DNA cytosine-N4 methyltransferase, SINEFUNGIN | | Authors: | Zhou, J, Horton, J.R, Cheng, X. | | Deposit date: | 2023-03-29 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Biochemical and structural characterization of the first-discovered metazoan DNA cytosine-N4 methyltransferase from the bdelloid rotifer Adineta vaga.
J.Biol.Chem., 299, 2023
|
|
7XLW
 
 | |
1QBJ
 
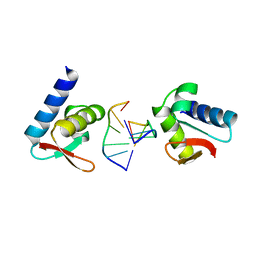 | | CRYSTAL STRUCTURE OF THE ZALPHA Z-DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*CP*G)-3'), PROTEIN (DOUBLE-STRANDED RNA SPECIFIC ADENOSINE DEAMINASE (ADAR1)) | | Authors: | Schwartz, T, Rould, M.A, Rich, A. | | Deposit date: | 1999-04-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA.
Science, 284, 1999
|
|
3PTA
 
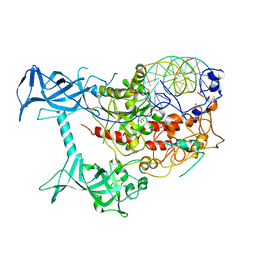 | | Crystal structure of human DNMT1(646-1600) in complex with DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*CP*GP*GP*AP*GP*GP*CP*TP*CP*AP*CP*GP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*CP*GP*TP*GP*AP*GP*CP*CP*TP*CP*CP*GP*CP*AP*GP*G)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
7Y01
 
 | |
2BOO
 
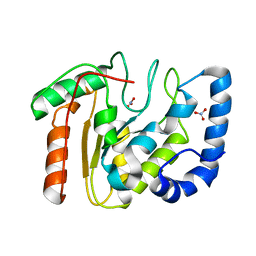 | | The crystal structure of Uracil-DNA N-Glycosylase (UNG) from Deinococcus radiodurans. | | Descriptor: | NITRATE ION, URACIL-DNA GLYCOSYLASE | | Authors: | Leiros, I, Moe, E, Smalas, A.O, McSweeney, S. | | Deposit date: | 2005-04-13 | | Release date: | 2005-07-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Uracil-DNA N-Glycosylase (Ung) from Deinococcus Radiodurans.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
151D
 
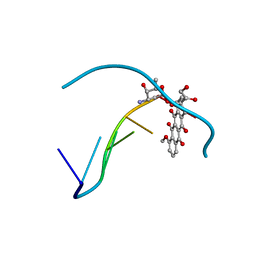 | | DIVERSITY OF WATER RING SIZE AT DNA INTERFACES: HYDRATION AND DYNAMICS OF DNA-ANTHRACYCLINE COMPLEXES | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), DOXORUBICIN | | Authors: | Lipscomb, L.A, Peek, M.E, Zhou, F.X, Bertrand, J.A, VanDerveer, D, Williams, L.D. | | Deposit date: | 1993-12-13 | | Release date: | 1994-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Water ring structure at DNA interfaces: hydration and dynamics of DNA-anthracycline complexes.
Biochemistry, 33, 1994
|
|
1D27
 
 | |
160D
 
 | |
1EOO
 
 | | ECORV BOUND TO COGNATE DNA | | Descriptor: | DNA (5'-D(*GP*AP*AP*GP*AP*TP*AP*TP*CP*TP*TP*C)-3'), TYPE II RESTRICTION ENZYME ECORV | | Authors: | Horton, N.C, Perona, J.J. | | Deposit date: | 2000-03-23 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystallographic snapshots along a protein-induced DNA-bending pathway.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1PJI
 
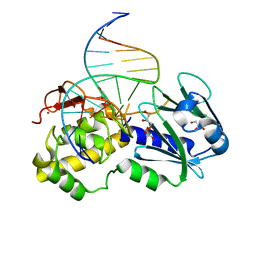 | | Crystal structure of wild type Lactococcus lactis FPG complexed to a 1,3 propanediol containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP*(PDI)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Pereira, K, Serre, L, Zelwer, C, Castaing, B. | | Deposit date: | 2003-06-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA.
Nucleic Acids Res., 33, 2005
|
|
