1MRE
 
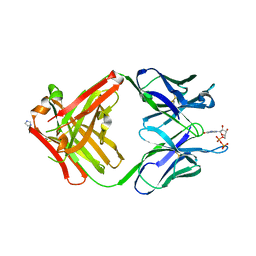 | | PREPARATION, CHARACTERIZATION AND CRYSTALLIZATION OF AN ANTIBODY FAB FRAGMENT THAT RECOGNIZES RNA. CRYSTAL STRUCTURES OF NATIVE FAB AND THREE FAB-MONONUCLEOTIDE COMPLEXES | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, IGG2B-KAPPA JEL103 FAB (HEAVY CHAIN), IGG2B-KAPPA JEL103 FAB (LIGHT CHAIN), ... | | Authors: | Pokkuluri, P.R, Cygler, M. | | Deposit date: | 1994-06-13 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Preparation, characterization and crystallization of an antibody Fab fragment that recognizes RNA. Crystal structures of native Fab and three Fab-mononucleotide complexes.
J.Mol.Biol., 243, 1994
|
|
3R8G
 
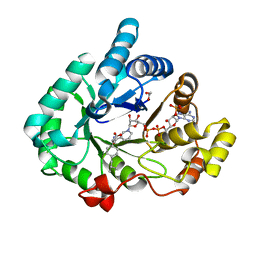 | | AKR1C3 complex with ibuprofen | | Descriptor: | (2R)-2-[4-(2-methylpropyl)phenyl]propanoic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C3, ... | | Authors: | Yosaatmadja, Y, Teague, R.M, Flanagan, J.U, Squire, C.J. | | Deposit date: | 2011-03-24 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structures of three classes of non-steroidal anti-inflammatory drugs in complex with aldo-keto reductase 1C3.
Plos One, 7, 2012
|
|
3BPK
 
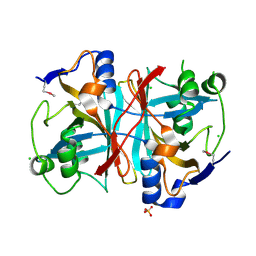 | | Crystal structure of nitrilotriacetate monooxygenase component B from Bacillus cereus | | Descriptor: | CHLORIDE ION, Nitrilotriacetate monooxygenase component B, SULFATE ION | | Authors: | Osipiuk, J, Quartey, P, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-01 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-ray crystal structure of nitrilotriacetate monooxygenase component B from Bacillus cereus.
To be Published
|
|
3RHH
 
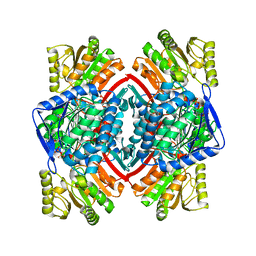 | | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP
To be Published
|
|
3RI8
 
 | | Xylanase C from Aspergillus kawachii D37N mutant | | Descriptor: | Endo-1,4-beta-xylanase 3 | | Authors: | Fushinobu, S, Uno, T, Kitaoka, M, Hayashi, K, Matsuzawa, H, Wakagi, T. | | Deposit date: | 2011-04-13 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational analysis of fungal family 11 xylanases on pH optimum determination
J.APPL.GLYOSCI., 58, 2011
|
|
3RIT
 
 | | Crystal structure of Dipeptide Epimerase from Methylococcus capsulatus complexed with Mg and dipeptide L-Arg-D-Lys | | Descriptor: | ARGININE, D-LYSINE, Dipeptide epimerase, ... | | Authors: | Lukk, T, Sakai, A, Song, L, Gerlt, J.A, Nair, S.K. | | Deposit date: | 2011-04-14 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RC3
 
 | | Human Mitochondrial Helicase Suv3 | | Descriptor: | ATP-dependent RNA helicase SUPV3L1, mitochondrial, AZIDE ION, ... | | Authors: | Dauter, Z, Jedrzejczak, R, Dauter, M, Szczesny, R, Stepien, P. | | Deposit date: | 2011-03-30 | | Release date: | 2011-05-11 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Human Suv3 protein reveals unique features among SF2 helicases.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3RCM
 
 | | crystal structure of EFI target 500140:TatD family hydrolase from Pseudomonas putida | | Descriptor: | ACETATE ION, CITRIC ACID, TatD family hydrolase, ... | | Authors: | Kim, J, Toro, R, Hillerich, B, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-20 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | crystal structure of EFI target 500140:TatD family hydrolase from Pseudomonas putida
TO BE PUBLISHED
|
|
6CCM
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 2-((3-bromobenzyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one | | Descriptor: | 2-{[(3-bromophenyl)methyl]amino}-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
6CCS
 
 | |
3RE7
 
 | | Copper (II) loaded Bullfrog Ferritin M chain | | Descriptor: | COPPER (II) ION, Ferritin, middle subunit | | Authors: | Bertini, I, Lalli, D, Mangani, S, Pozzi, C, Rosa, C, Turano, P. | | Deposit date: | 2011-04-02 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural insights into the ferroxidase site of ferritins from higher eukaryotes.
J.Am.Chem.Soc., 134, 2012
|
|
3RJY
 
 | | Crystal Structure of Hyperthermophilic Endo-beta-1,4-glucanase in complex with substrate | | Descriptor: | Endoglucanase FnCel5A, PHOSPHATE ION, alpha-D-glucopyranose | | Authors: | Zheng, B, Yang, W, Zhao, X, Wang, Y, Lou, Z, Rao, Z, Feng, Y. | | Deposit date: | 2011-04-15 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of hyperthermophilic Endo-beta-1,4-glucanase: Implications for catalytic mechanism and thermostability.
J.Biol.Chem., 287, 2012
|
|
3QK5
 
 | | Crystal structure of fatty acid amide hydrolase with small molecule inhibitor | | Descriptor: | (3-{(3R)-1-[4-(1-benzothiophen-2-yl)pyrimidin-2-yl]piperidin-3-yl}-2-methyl-1H-pyrrolo[2,3-b]pyridin-1-yl)acetonitrile, 1,2-ETHANEDIOL, Fatty-acid amide hydrolase 1, ... | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-01-31 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of potent, noncovalent fatty acid amide hydrolase (FAAH) inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3C82
 
 | | Bacteriophage lysozyme T4 lysozyme mutant K85A/R96H | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3QJL
 
 | |
3Q93
 
 | | Crystal Structure of Human 8-oxo-dGTPase (MTH1) | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Tresaugues, L, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Thorsell, A.G, Van Der Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human 8-oxo-dGTPase (MTH1)
To be Published
|
|
3BLV
 
 | | Yeast Isocitrate Dehydrogenase with Citrate Bound in the Regulatory Subunits | | Descriptor: | CITRATE ANION, Isocitrate dehydrogenase [NAD] subunit 1, Isocitrate dehydrogenase [NAD] subunit 2 | | Authors: | Taylor, A.B, Hu, G, Hart, P.J, McAlister-Henn, L. | | Deposit date: | 2007-12-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Allosteric Motions in Structures of Yeast NAD+-specific Isocitrate Dehydrogenase.
J.Biol.Chem., 283, 2008
|
|
3BNC
 
 | | Lipoxygenase-1 (Soybean) I553G Mutant | | Descriptor: | ACETIC ACID, FE (III) ION, Seed lipoxygenase-1 | | Authors: | Tomchick, D.R. | | Deposit date: | 2007-12-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Enzyme structure and dynamics affect hydrogen tunneling: the impact of a remote side chain (I553) in soybean lipoxygenase-1.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C5C
 
 | | Crystal structure of human Ras-like, family 12 protein in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAS-like protein 12, ... | | Authors: | Shen, L, Tong, Y, Tempel, W, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of human Ras-like, family 12 protein in complex with GDP.
To be Published
|
|
1MUP
 
 | | PHEROMONE BINDING TO TWO RODENT URINARY PROTEINS REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2-(SEC-BUTYL)THIAZOLE, CADMIUM ION, MAJOR URINARY PROTEIN | | Authors: | Bocskei, Z, Flower, D.R, Groom, C.R, Phillips, S.E.V, North, A.C.T. | | Deposit date: | 1992-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Pheromone binding to two rodent urinary proteins revealed by X-ray crystallography.
Nature, 360, 1992
|
|
6CCO
 
 | |
3C7Y
 
 | | Mutant R96A OF T4 lysozyme in wildtype background at 298K | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, lysozyme | | Authors: | Mooers, B.H.M. | | Deposit date: | 2008-02-08 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Contributions of all 20 amino acids at site 96 to the stability and structure of T4 lysozyme.
Protein Sci., 18, 2009
|
|
3QNG
 
 | |
1N57
 
 | | Crystal Structure of Chaperone Hsp31 | | Descriptor: | Chaperone Hsp31, MAGNESIUM ION | | Authors: | Quigley, P.M, Korotkov, K, Baneyx, F, Hol, W.G.J. | | Deposit date: | 2002-11-04 | | Release date: | 2003-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6A Crystal Structure of the Class of Chaperone Represented by
Escherichia coli Hsp31 Reveals a Putative Catalytic Triad
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6CCQ
 
 | |
