1AH7
 
 | | PHOSPHOLIPASE C FROM BACILLUS CEREUS | | Descriptor: | PHOSPHOLIPASE C, ZINC ION | | Authors: | Greaves, R. | | Deposit date: | 1997-04-14 | | Release date: | 1997-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | High-resolution (1.5 A) crystal structure of phospholipase C from Bacillus cereus.
Nature, 338, 1989
|
|
2HUC
 
 | |
1P5X
 
 | | STRUCTURE OF THE D55N MUTANT OF PHOSPHOLIPASE C FROM BACILLUS CEREUS | | Descriptor: | Phospholipase C, ZINC ION | | Authors: | Antikainen, N.M, Monzingo, A.F, Franklin, C.L, Robertus, J.D, Martin, S.F. | | Deposit date: | 2003-04-28 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Using X-ray crystallography of the Asp55Asn mutant of the phosphatidylcholine-preferring phospholipase C from Bacillus cereus to support the mechanistic role of Asp55 as the general base.
Arch.Biochem.Biophys., 417, 2003
|
|
1P6D
 
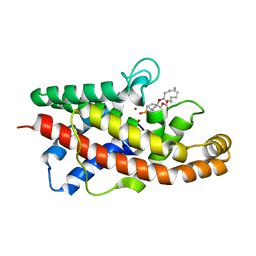 | | STRUCTURE OF THE D55N MUTANT OF PHOSPHOLIPASE C FROM BACILLUS CEREUS IN COMPLEX WITH (3S)-3,4,DI-N-HEXANOYLOXYBUTYL-1-PHOSPHOCHOLINE | | Descriptor: | (3S)-3,4-DI-N-HEXANOYLOXYBUTYL-1-PHOSPHOCHOLINE, PHOSPHOLIPASE C, ZINC ION | | Authors: | Antikainen, N.M, Monzingo, A.F, Franklin, C.L, Robertus, J.D, Martin, S.F. | | Deposit date: | 2003-04-29 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Using X-ray crystallography of the Asp55Asn mutant of the phosphatidylcholine-preferring phospholipase C from Bacillus cereus to support the mechanistic role of Asp55 as the general base.
Arch.Biochem.Biophys., 417, 2003
|
|
2FGN
 
 | |
2FFZ
 
 | |
1P6E
 
 | | STRUCTURE OF THE D55N MUTANT OF PHOSPHOLIPASE C FROM BACILLUS CEREUS IN COMPLEX WITH 1,2-DI-N-PENTANOYL-SN-GLYCERO-3-DITHIOPHOSPHOCHOLINE | | Descriptor: | 1,2-DI-N-PENTANOYL-SN-GLYCERO-3-DITHIOPHOSPHOCHOLINE, Phospholipase C, ZINC ION | | Authors: | Antikainen, N.M, Monzingo, A.F, Franklin, C.L, Robertus, J.D, Martin, S.F. | | Deposit date: | 2003-04-29 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Using X-ray crystallography of the Asp55Asn mutant of the phosphatidylcholine-preferring phospholipase C from Bacillus cereus to support the mechanistic role of Asp55 as the general base.
Arch.Biochem.Biophys., 417, 2003
|
|
