1C6P
 
 | |
1OWZ
 
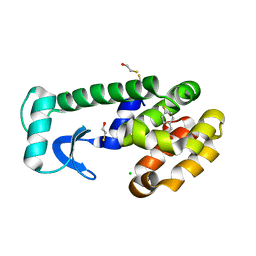 | | T4 Lysozyme Cavity Mutant L99A/M102Q Bound With 4-FluoroPhenEthyl Alcohol | | Descriptor: | 4-FLUOROPHENETHYL ALCOHOL, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2003-03-31 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Testing a Flexible-receptor Docking Algorithm in a Model Binding Site
J.Mol.Biol., 337, 2004
|
|
1C6H
 
 | |
1C6G
 
 | |
1C6Q
 
 | |
1C6I
 
 | |
1C6J
 
 | |
1C6K
 
 | |
1C6L
 
 | |
1JTM
 
 | |
1PQJ
 
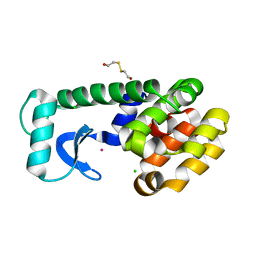 | | T4 LYSOZYME CORE REPACKING MUTANT A111V/CORE10/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | REPACKING THE CORE OF T4 LYSOZYME BY AUTOMATED DESIGN
J.Mol.Biol., 332, 2003
|
|
1CV4
 
 | | T4 LYSOZYME MUTANT L118M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1QUO
 
 | | L99A/E108V MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | Authors: | Wray, J, Baase, W.A, Lindstrom, J.D, Poteete, A.R, Matthews, B.W. | | Deposit date: | 1999-07-01 | | Release date: | 1999-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of a non-contiguous second-site revertant in T4 lysozyme shows that increasing the rigidity of a protein can enhance its stability.
J.Mol.Biol., 292, 1999
|
|
1CV6
 
 | | T4 LYSOZYME MUTANT V149M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-22 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1L00
 
 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | Deposit date: | 1992-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
225L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, PARA-XYLENE, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
233L
 
 | | T4 LYSOZYME MUTANT M120L | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-07 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
1L28
 
 | |
1L55
 
 | |
1L69
 
 | |
1LGU
 
 | | T4 Lysozyme Mutant L99A/M102Q | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
223L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BENZENE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
1L79
 
 | |
1L84
 
 | |
1L34
 
 | |
