4FK7
 
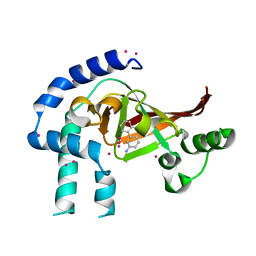 | | Crystal structure of Certhrax catalytic domain | | Descriptor: | CHLORIDE ION, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Putative ADP-ribosyltransferase Certhrax, ... | | Authors: | Hong, B.S, Dimov, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-12 | | Release date: | 2012-09-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Certhrax Toxin, an Anthrax-related ADP-ribosyltransferase from Bacillus cereus.
J.Biol.Chem., 287, 2012
|
|
3NTS
 
 | |
1UZI
 
 | | C3 EXOENZYME FROM CLOSTRIDIUM BOTULINUM, TETRAGONAL FORM | | Descriptor: | CYCLO-TETRAMETAVANADATE, GLYCEROL, MONO-ADP-RIBOSYLTRANSFERASE C3, ... | | Authors: | Evans, H.R, Holloway, D.E, Sutton, J.M, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | C3 Exoenzyme from Clostridium Botulinum: Structure of a Tetragonal Crystal Form and a Reassessment of Nad-Induced Flexure
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5URP
 
 | | Plx2a, an ADP-ribosyltransferase toxin from Paenibacillus larvae | | Descriptor: | GLYCEROL, Toxin 2A | | Authors: | Ravulapalli, R, Heney, K, Ebeling, J, Genersch, E, Merrill, A.R. | | Deposit date: | 2017-02-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional characterization of Plx2a, an ADP-ribosyltransferase toxin from Paenibacillus larvae
Environ.Microbiol., 2017
|
|
3U0J
 
 | |
4FML
 
 | |
5WTZ
 
 | | Crystal structure of C. perfringens iota-like enterotoxin CPILE-a with NAD+ | | Descriptor: | Binary enterotoxin of Clostridium perfringens component a, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Toniti, W, Yoshida, T, Tsurumura, T, Irikura, D, Tsuge, H. | | Deposit date: | 2016-12-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin
PLoS ONE, 12, 2017
|
|
5WU0
 
 | | Crystal structure of C. perfringens iota-like enterotoxin CPILE-a with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Toniti, W, Yoshida, T, Tsurumura, T, Irikura, D, Tsuge, H. | | Deposit date: | 2016-12-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin
PLoS ONE, 12, 2017
|
|
3BW8
 
 | | Crystal structure of the Clostridium limosum C3 exoenzyme | | Descriptor: | Mono-ADP-ribosyltransferase C3, SULFATE ION | | Authors: | Vogelsgesang, M, Stieglitz, B, Herrmann, C, Pautsch, A, Aktories, K. | | Deposit date: | 2008-01-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Clostridium limosum C3 exoenzyme.
Febs Lett., 582, 2008
|
|
5H03
 
 | | Crystal structure of an ADP-ribosylating toxin BECa from C. perfringens | | Descriptor: | Binary enterotoxin of Clostridium perfringens component a | | Authors: | Kawahara, K, Yonogi, S, Munetomo, R, Oki, H, Yoshida, T, Ohkubo, T, Kumeda, Y, Matsuda, S, Kodama, T, Iida, T, Nakamura, S. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of the ADP-ribosylating component of BEC, the binary enterotoxin of Clostridium perfringens.
Biochem.Biophys.Res.Commun., 480, 2016
|
|
2C8B
 
 | | Structure of the ARTT motif Q212A mutant C3bot1 Exoenzyme (Free state, crystal form II) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
2C8G
 
 | | Structure of the PN loop Q182A mutant C3bot1 Exoenzyme (Free state, crystal form I) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Properties of Wild-Type and Two Artt Motif Mutants Clostridium Botulinum C3 Exoenzyme Isoform 1 in Different Substrate Complexed States and Crystal Forms.
To be Published
|
|
5GTT
 
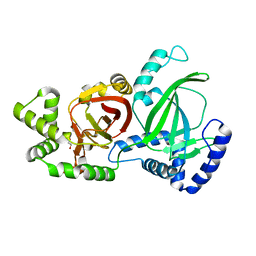 | | Crystal structure of C. perfringens iota-like enterotoxin CPILE-a | | Descriptor: | 1,2-ETHANEDIOL, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Toniti, W, Yoshida, T, Tsurumura, T, Irikura, D, Tsuge, H. | | Deposit date: | 2016-08-23 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Crystal structure and structure-based mutagenesis of actin-specific ADP-ribosylating toxin CPILE-a as novel enterotoxin
PLoS ONE, 12, 2017
|
|
5H04
 
 | | Crystal structure of an ADP-ribosylating toxin BECa of a novel binary enterotoxin of C. perfringens with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Binary enterotoxin of Clostridium perfringens component a | | Authors: | Kawahara, K, Yonogi, S, Munetomo, R, Oki, H, Yoshida, T, Ohkubo, T, Kumeda, Y, Matsuda, S, Kodama, T, Iida, T, Nakamura, S. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | Crystal structure of the ADP-ribosylating component of BEC, the binary enterotoxin of Clostridium perfringens.
Biochem.Biophys.Res.Commun., 480, 2016
|
|
2GWL
 
 | |
2J3V
 
 | | Crystal structure of the enzymatic component C2-I of the C2-toxin from Clostridium botulinum at pH 3.0 | | Descriptor: | C2 TOXIN COMPONENT I, GLYCEROL, SULFATE ION | | Authors: | Schleberger, C, Hochmann, H, Barth, H, Aktories, K, Schulz, G.E. | | Deposit date: | 2006-08-23 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure and Action of the Binary C2 Toxin from Clostridium Botulinum.
J.Mol.Biol., 364, 2006
|
|
2J3X
 
 | | Crystal structure of the enzymatic component C2-I of the C2-toxin from Clostridium botulinum at pH 3.0 (mut-S361R) | | Descriptor: | C2 TOXIN COMPONENT I, GLYCEROL, SULFATE ION | | Authors: | Schleberger, C, Hochmann, H, Barth, H, Aktories, K, Schulz, G.E. | | Deposit date: | 2006-08-23 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Action of the Binary C2 Toxin from Clostridium Botulinum.
J.Mol.Biol., 364, 2006
|
|
2J3Z
 
 | | Crystal structure of the enzymatic component C2-I of the C2-toxin from Clostridium botulinum at pH 6.1 | | Descriptor: | C2 TOXIN COMPONENT I, COBALT (II) ION, GLYCEROL, ... | | Authors: | Schleberger, C, Hochmann, H, Barth, H, Aktories, K, Schulz, G.E. | | Deposit date: | 2006-08-23 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Action of the Binary C2 Toxin from Clostridium Botulinum.
J.Mol.Biol., 364, 2006
|
|
2GWM
 
 | |
6X41
 
 | |
6X6W
 
 | |
6X6X
 
 | |
6X6V
 
 | |
6X6R
 
 | | Crystal structure of C.difficile ribosyltransferase CDTa in complex with pCl-phenylthioDADMeImmA | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-{[(4-chlorophenyl)sulfanyl]methyl}pyrrolidin-3-ol, ADP-ribosyltransferase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pozharski, E. | | Deposit date: | 2020-05-29 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of C.difficile ribosyltransferase CDTa in complex with pCl-phenylthioDADMeImmA
To Be Published
|
|
2WN7
 
 | | Structural Basis for Substrate Recognition in the Enzymatic Component of ADP-ribosyltransferase Toxin CDTa from Clostridium difficile | | Descriptor: | ADP-RIBOSYLTRANSFERASE ENZYMATIC COMPONENT, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sundriyal, A, Roberts, A.K, Shone, C.C, Acharya, K.R. | | Deposit date: | 2009-07-07 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis for Substrate Recognition in the Enzymatic Component of Adp-Ribosyltransferase Toxin Cdta from Clostridium Difficile.
J.Biol.Chem., 284, 2009
|
|
