6N0W
 
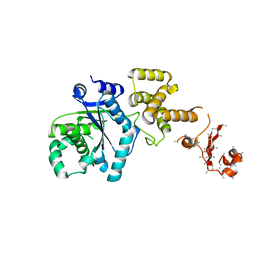 | |
6N2J
 
 | | Tetrahydropyridopyrimidines as Covalent Inhibitors of KRAS-G12C | | Descriptor: | 1-{4-[7-(naphthalen-1-yl)-5,6,7,8-tetrahydropyrido[3,4-d]pyrimidin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Vigers, G.P. | | Deposit date: | 2018-11-13 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Tetrahydropyridopyrimidines as Irreversible Covalent Inhibitors of KRAS-G12C with In Vivo Activity.
ACS Med Chem Lett, 9, 2018
|
|
7BSV
 
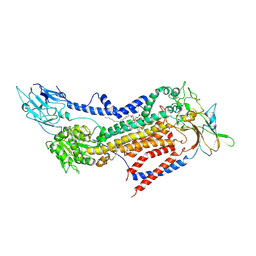 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in PtdSer-occluded E2-AlF state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, CDC50A, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7A1H
 
 | | Crystal structure of wild-type CI2 | | Descriptor: | SULFATE ION, Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Olsen, J.G, Teilum, K, Hamborg, L, Roche, J.V. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synergistic stabilization of a double mutant in chymotrypsin inhibitor 2 from a library screen in E. coli.
Commun Biol, 4, 2021
|
|
7AE3
 
 | | Cryo-EM structure of human RNA Polymerase III elongation complex 3 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7AEA
 
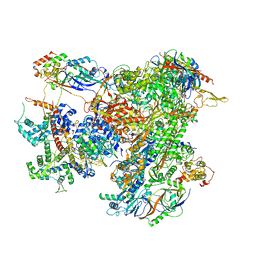 | | Cryo-EM structure of human RNA Polymerase III elongation complex 2 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZTO
 
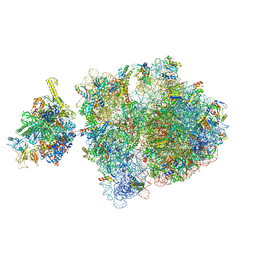 | | E. coli 70S-RNAP expressome complex in uncoupled state 1 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
7AE1
 
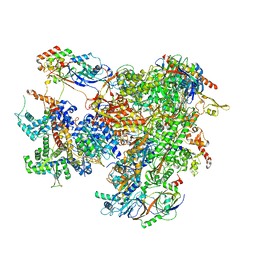 | | Cryo-EM structure of human RNA Polymerase III elongation complex 1 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZRU
 
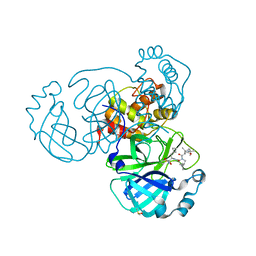 | | Crystal structure of SARS CoV2 main protease in complex with inhibitor Boceprevir | | Descriptor: | DIMETHYL SULFOXIDE, Main Protease, boceprevir (bound form) | | Authors: | Oerlemans, R, Wang, W, Lunev, S, Domling, A, Groves, M.R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Repurposing the HCV NS3-4A protease drug boceprevir as COVID-19 therapeutics.
Rsc Med Chem, 12, 2020
|
|
2FQ9
 
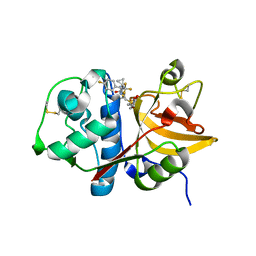 | | Cathepsin S with nitrile inhibitor | | Descriptor: | N-(1-CYANOCYCLOPROPYL)-3-({[(2S)-5-OXOPYRROLIDIN-2-YL]METHYL}SULFONYL)-N~2~-[(1S)-2,2,2-TRIFLUORO-1-(4-FLUOROPHENYL)ETHYL]-L-ALANINAMIDE, cathepsin S | | Authors: | Somoza, J.R. | | Deposit date: | 2006-01-17 | | Release date: | 2006-03-21 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cathepsin S with nitrile inhibitor
To be Published
|
|
6NLI
 
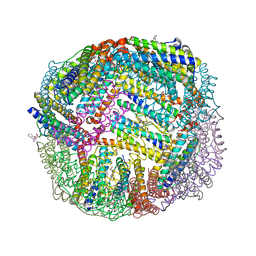 | | 1.90 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor (analog 11) | | Descriptor: | 4-{[(2-hydroxyphenyl)methyl]amino}-1H-isoindole-1,3(2H)-dione, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Punchi-Hewage, A, Battaile, K.P, Yao, H, Nammalwar, B, Gnanasekaran, K.K, Bunce, R.A, Reitz, A.B, Rivera, M. | | Deposit date: | 2019-01-08 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Small Molecule Inhibitors of the BfrB-Bfd Interaction Decrease Pseudomonas aeruginosa Fitness and Potentiate Fluoroquinolone Activity.
J.Am.Chem.Soc., 141, 2019
|
|
6SD5
 
 | | Structure of the RBM2 inner ring of Salmonella flagella MS-ring protein FliF with 22-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6NOT
 
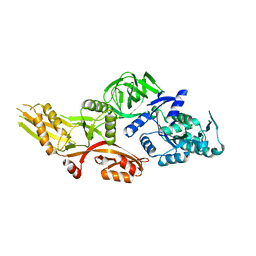 | |
2FZF
 
 | | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Fu, Z.-Q, Liu, Z.-J, Lee, D, Kelley, L, Chen, L, Tempel, W, Shah, N, Horanyi, P, Lee, H.S, Habel, J, Dillard, B.D, Nguyen, D, Chang, S.-H, Zhang, H, Chang, J, Sugar, F.J, Poole, F.L, Jenney Jr, F.E, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hypothetical Protein Pfu-1136390-001 From Pyrococcus furiosus
To be published
|
|
6NRH
 
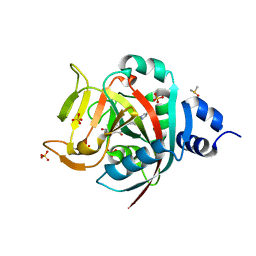 | | Crystal Structure of human PARP-1 ART domain bound inhibitor UTT63 | | Descriptor: | 3-hydroxy-2-({4-[4-(pyrimidin-2-yl)piperazine-1-carbonyl]phenyl}methyl)-1-benzofuran-7-carboxamide, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Langelier, M.F, Pascal, J.M. | | Deposit date: | 2019-01-23 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design and Synthesis of Poly(ADP-ribose) Polymerase Inhibitors: Impact of Adenosine Pocket-Binding Motif Appendage to the 3-Oxo-2,3-dihydrobenzofuran-7-carboxamide on Potency and Selectivity.
J.Med.Chem., 62, 2019
|
|
7BSW
 
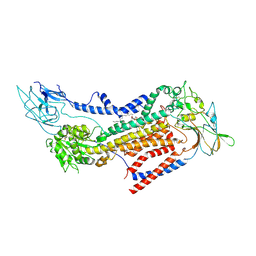 | | Cryo-EM structure of a human ATP11C-CDC50A flippase in PtdEtn-occluded E2-AlF state | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ATP11C, ... | | Authors: | Abe, K, Nishizawa, T, Nakanishi, H. | | Deposit date: | 2020-03-31 | | Release date: | 2020-09-30 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Transport Cycle of Plasma Membrane Flippase ATP11C by Cryo-EM.
Cell Rep, 32, 2020
|
|
7BT7
 
 | | F-actin-ADP complex structure | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Kumari, A, Ragunath, V.K, Sirajuddin, M. | | Deposit date: | 2020-03-31 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into actin filament recognition by commonly used cellular actin markers.
Embo J., 39, 2020
|
|
6N9E
 
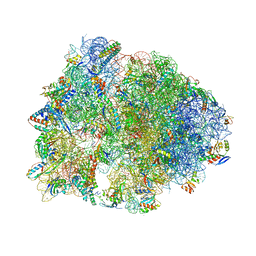 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with a short substrate mimic CC-Pmn and bound to mRNA and P-site tRNA at 3.7A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S19, ... | | Authors: | Melnikov, S.V, Khabibullina, N.F, Mairhofer, E, Vargas-Rodriguez, O, Reynolds, N.M, Micura, R, Soll, D, Polikanov, Y.S. | | Deposit date: | 2018-12-03 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanistic insights into the slow peptide bond formation with D-amino acids in the ribosomal active site.
Nucleic Acids Res., 47, 2019
|
|
2FVU
 
 | | Structure of the yeast Sir3 BAH domain | | Descriptor: | Regulatory protein SIR3 | | Authors: | Xu, R.M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain.
Mol.Cell.Biol., 26, 2006
|
|
7BXT
 
 | | The cryo-EM structure of CENP-A nucleosome in complex with CENP-C peptide and CENP-N N-terminal domain | | Descriptor: | CENP-C, DNA (145-mer), Histone H2A type 1-B/E, ... | | Authors: | Ariyoshi, M, Makino, F, Fukagawa, T. | | Deposit date: | 2020-04-20 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Embo J., 40, 2021
|
|
6T8A
 
 | | Thrombin in complex with diphenyl ((4-carbamimidoylphenyl)((S)-1-((R)-3-cyclohexyl 2-((phenylmethyl)sulfonamido)propanoyl)pyrrolidine-2-carboxamido)methyl)phosphonate (MI-492) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Hirudin variant-2, ... | | Authors: | Ngaha, S.A, Sandner, A, Huber, S, Heine, A, Steinmetzer, T, Pilgram, O. | | Deposit date: | 2019-10-24 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Thrombin in complex with diphenyl ((4-carbamimidoylphenyl)((S)-1-((R)-3-cyclohexyl-2-((phenylmethyl)sulfonamido)propanoyl)pyrrolidine-2-carboxamido)methyl)phosphonate (MI-492)
to be published
|
|
6TF9
 
 | | Structure of the vertebrate gamma-Tubulin Ring Complex | | Descriptor: | Actin, cytoplasmic 1, Belt helices 1,2,3,4, ... | | Authors: | Zupa, E, Pfeffer, S. | | Deposit date: | 2019-11-13 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Insights into the assembly and activation of the microtubule nucleator gamma-TuRC.
Nature, 578, 2020
|
|
6YHS
 
 | | Acinetobacter baumannii ribosome-amikacin complex - 50S subunit | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-03-30 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
6YW5
 
 | | The structure of the small subunit of the mitoribosome from Neurospora crassa | | Descriptor: | 16S rRNA, 3-hydroxyisobutyryl-CoA hydrolase, mitochondrial, ... | | Authors: | Amunts, A, Itoh, Y, Naschberger, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi.
Nat Commun, 11, 2020
|
|
6NLY
 
 | |
