1UTL
 
 | | Trypsin specificity as elucidated by LIE calculations, X-ray structures and association constant measurements | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-PHENYLPROPYLAMINE, CALCIUM ION, ... | | Authors: | Leiros, H.-K.S, Brandsdal, B.O, Andersen, O.A, Os, V, Leiros, I, Helland, R, Otlewski, J, Willassen, N.P, Smalas, A.O. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Trypsin Specificity as Elucidated by Lie Calculations, X-Ray Structures, and Association Constant Measurements
Protein Sci., 13, 2004
|
|
1YAL
 
 | | CARICA PAPAYA CHYMOPAPAIN AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | CHYMOPAPAIN | | Authors: | Maes, D, Bouckaert, J, Poortmans, F, Wyns, L, Looze, Y. | | Deposit date: | 1996-06-20 | | Release date: | 1996-12-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of chymopapain at 1.7 A resolution.
Biochemistry, 35, 1996
|
|
1VBC
 
 | | POLIOVIRUS (TYPE 3, SABIN STRAIN) (P3/SABIN, P3/LEON/12A(1)B) COMPLEXED WITH R77975 | | Descriptor: | (METHYLPYRIDAZINE PIPERIDINE ETHYLOXYPHENYL)ETHYLACETATE, MYRISTIC ACID, POLIOVIRUS TYPE 3 | | Authors: | Grant, R.A, Hiremath, C.N, Filman, D.J, Syed, R, Andries, K, Hogle, J.M. | | Deposit date: | 1996-01-02 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design.
Curr.Biol., 4, 1994
|
|
1Y37
 
 | |
1UUE
 
 | | a-SPECTRIN SH3 DOMAIN (V44T, D48G MUTANT) | | Descriptor: | SPECTRIN ALPHA CHAIN | | Authors: | Vega, M.C, Fernandez, A, Wilmanns, M, Serrano, L. | | Deposit date: | 2003-12-18 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Solvation in Protein Folding Analysis: Combination of Theoretical and Experimental Approaches
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1UV0
 
 | | Pancreatitis-associated protein 1 from human | | Descriptor: | PANCREATITIS-ASSOCIATED PROTEIN 1, ZINC ION | | Authors: | Abergel, C, Shepard, W, Christal, L. | | Deposit date: | 2004-01-12 | | Release date: | 2004-01-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystallization and preliminary crystallographic study of HIP/PAP, a human C-lectin overexpressed in primary liver cancers.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1Y4I
 
 | | Crystal structure of Citrobacter Freundii L-methionine-lyase | | Descriptor: | SULFATE ION, methionine gamma-lyase | | Authors: | Revtovich, S.V, Mamaeva, D.V, Morozova, E.A, Nikulin, A.D, Nikonov, S.V, Garber, M.B, Demidkina, T.V. | | Deposit date: | 2004-12-01 | | Release date: | 2005-06-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Citrobacter freundii L-methionine gamma-lyase.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
1UZE
 
 | | Complex of the anti-hypertensive drug enalaprilat and the human testicular angiotensin I-converting enzyme | | Descriptor: | 1-((2S)-2-{[(1S)-1-CARBOXY-3-PHENYLPROPYL]AMINO}PROPANOYL)-L-PROLINE, ANGIOTENSIN CONVERTING ENZYME, CHLORIDE ION, ... | | Authors: | Natesh, R, Schwager, S.L.U, Evans, H.R, Sturrock, E.D, Acharya, K.R. | | Deposit date: | 2004-03-11 | | Release date: | 2004-07-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Details on the Binding of Antihypertensive Drugs Captopril and Enalaprilat to Human Testicular Angiotensin I-Converting Enzyme
Biochemistry, 43, 2004
|
|
1YD8
 
 | | COMPLEX OF HUMAN GGA3 GAT DOMAIN AND UBIQUITIN | | Descriptor: | ADP-RIBOSYLATION FACTOR BINDING PROTEIN GGA3, UBIQUIN | | Authors: | Prag, G, Lee, S, Mattera, R, Arighi, C.N, Beach, B.M, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanism for ubiquitinated-cargo recognition by the Golgi-localized, {gamma}-ear-containing, ADP-ribosylation-factor-binding proteins
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1V00
 
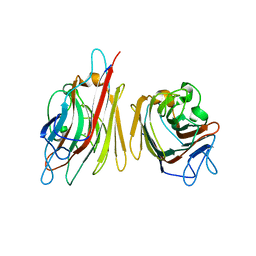 | | Erythrina cristagalli lectin | | Descriptor: | CALCIUM ION, LECTIN (ECL), MANGANESE (II) ION, ... | | Authors: | Turton, K, Natesh, R, Thiyagarajan, N, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2004-03-21 | | Release date: | 2004-06-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Erythrina Cristagalli Lectin with Bound N-Linked Oligosaccharide and Lactose
Glycobiology, 14, 2004
|
|
1UWY
 
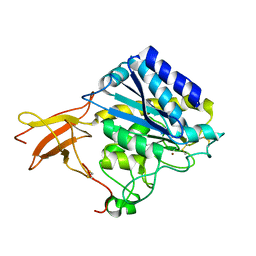 | | Crystal structure of human carboxypeptidase M | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBOXYPEPTIDASE M, ZINC ION | | Authors: | Maskos, K, Reverter, D, Bode, W. | | Deposit date: | 2004-02-17 | | Release date: | 2004-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Human Carboxypeptidase M, a Membrane-Bound Enzyme that Regulates Peptide Hormone Activity
J.Mol.Biol., 338, 2004
|
|
1YAC
 
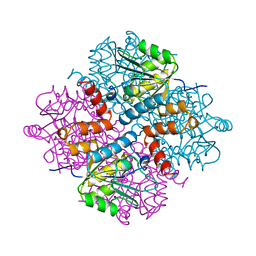 | |
2EA9
 
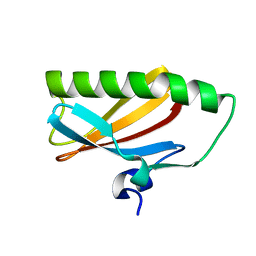 | |
2EAU
 
 | | Crystal structure of the SR CA2+-ATPASE with bound CPA in the presence of curcumin | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, PHOSPHATIDYLETHANOLAMINE, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2007-02-02 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1V3D
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III: complex with NEU5AC2EN | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1YFY
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase from Ralstonia metallidurans complexed with 3-hydroxyanthranilic acid | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate-3,4-dioxygenase, ... | | Authors: | Zhang, Y, Colabroy, K.L, Begley, T.P, Ealick, S.E. | | Deposit date: | 2005-01-04 | | Release date: | 2005-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Studies on 3-Hydroxyanthranilate-3,4-dioxygenase: The Catalytic Mechanism of a Complex Oxidation Involved in NAD Biosynthesis.
Biochemistry, 44, 2005
|
|
1Y7E
 
 | |
1Y5N
 
 | | The crystal structure of the NarGHI mutant NarI-K86A in complex with pentachlorophenol | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1V30
 
 | | Crystal Structure Of Uncharacterized Protein PH0828 From Pyrococcus horikoshii | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Hypothetical UPF0131 protein PH0828 | | Authors: | Tajika, Y, Sakai, N, Yao, M, Watanabe, N, Tanaka, I. | | Deposit date: | 2003-10-21 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of hypothetical protein PH0828 from Pyrococcus horikoshii.
Proteins, 57, 2004
|
|
1GL2
 
 | | Crystal structure of an endosomal SNARE core complex | | Descriptor: | ENDOBREVIN, SYNTAXIN 7, SYNTAXIN 8, ... | | Authors: | Antonin, W, Becker, S, Jahn, R, Schneider, T.R. | | Deposit date: | 2001-08-22 | | Release date: | 2002-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Endosomal Snare Complex Reveals Common Structural Principles of All Snares.
Nat.Struct.Biol., 9, 2001
|
|
2EEV
 
 | | Guanine riboswitch U22C, A52G mutant bound to hypoxanthine | | Descriptor: | ACETATE ION, COBALT HEXAMMINE(III), HYPOXANTHINE, ... | | Authors: | Gilbert, S.D, Love, C.E, Batey, R.T. | | Deposit date: | 2007-02-19 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mutational analysis of the purine riboswitch aptamer domain
Biochemistry, 46, 2007
|
|
1YA9
 
 | | Crystal Structure of the 22kDa N-Terminal Fragment of Mouse Apolipoprotein E | | Descriptor: | Apolipoprotein E | | Authors: | Peters-Libeu, C.A, Rutenber, E, Newhouse, Y, Hatters, D.M, Weisgraber, K.H. | | Deposit date: | 2004-12-17 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Engineering conformational destabilization into mouse apolipoprotein E. A model for a unique property of human apolipoprotein E4
J.Biol.Chem., 280, 2005
|
|
2EG2
 
 | | The crystal structure of PII protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Nitrogen regulatory protein P-II | | Authors: | Sakai, H, Shinkai, A, Kitamura, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-27 | | Release date: | 2008-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of PII protein
To be Published
|
|
1V7C
 
 | |
2E7Y
 
 | | High resolution structure of T. maritima tRNase Z | | Descriptor: | S-1,2-PROPANEDIOL, SULFATE ION, ZINC ION, ... | | Authors: | Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-15 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the flexible arm of Thermotoga maritima tRNase Z differs from those of homologous enzymes
Acta Crystallogr.,Sect.F, 63, 2007
|
|
