2KP5
 
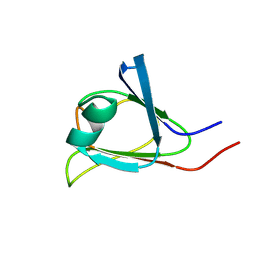 | |
1KOS
 
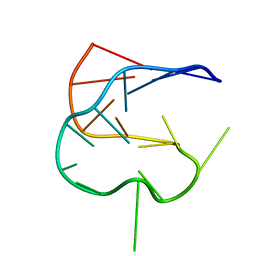 | | SOLUTION NMR STRUCTURE OF AN ANALOG OF THE YEAST TRNA PHE T STEM LOOP CONTAINING RIBOTHYMIDINE AT ITS NATURALLY OCCURRING POSITION | | Descriptor: | 5'-R(*CP*UP*GP*UP*GP*(5MU)P*UP*CP*GP*AP*UP*(CH)P*CP*AP*CP*AP*G)- 3' | | Authors: | Koshlap, K.M, Guenther, R, Sochacka, E, Malkiewicz, A, Agris, P.F. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A distinctive RNA fold: the solution structure of an analogue of the yeast tRNAPhe T Psi C domain.
Biochemistry, 38, 1999
|
|
2MTD
 
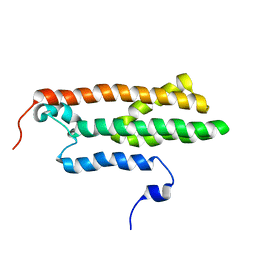 | |
1YI2
 
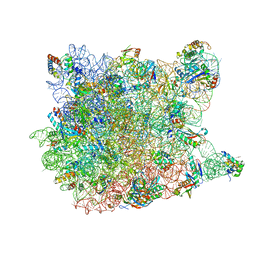 | | Crystal Structure Of Erythromycin Bound To The G2099A Mutant 50S Ribosomal Subunit Of Haloarcula Marismortui | | Descriptor: | 23S Ribosomal RNA, 50S RIBOSOMAL PROTEIN L10E, 50S RIBOSOMAL PROTEIN L11P, ... | | Authors: | Tu, D, Blaha, G, Moore, P.B, Steitz, T.A. | | Deposit date: | 2005-01-11 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance.
Cell(Cambridge,Mass.), 121, 2005
|
|
1GEN
 
 | | C-TERMINAL DOMAIN OF GELATINASE A | | Descriptor: | CALCIUM ION, CHLORIDE ION, GELATINASE A, ... | | Authors: | Libson, A.M, Gittis, A.G, Collier, I.E, Marmer, B.L, Goldberg, G.G, Lattman, E.E. | | Deposit date: | 1995-07-19 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the haemopexin-like C-terminal domain of gelatinase A.
Nat.Struct.Biol., 2, 1995
|
|
2HWN
 
 | |
2K6U
 
 | | The Solution Structure of a Conformationally Restricted Fully Active Derivative of the Human Relaxin-like Factor (RLF) | | Descriptor: | Insulin-like 3 A chain, Insulin-like 3 B chain | | Authors: | Bullesbach, E.E, Hass, M.A.S, Jensen, M.R, Hansen, D.F, Kristensen, S.M, Schwabe, C, Led, J.J. | | Deposit date: | 2008-07-24 | | Release date: | 2008-12-16 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a conformationally restricted fully active derivative of the human relaxin-like factor
Biochemistry, 47, 2008
|
|
1LUH
 
 | | SOLUTION NMR STRUCTURE OF SELF-COMPLIMENTARY DUPLEX 5'-D(TCCG*CGGA)2 CONTAINING A TRIMETHYLENE CROSSLINK AT THE N2 POSITION OF G* | | Descriptor: | 5'-D(*TP*CP*CP*(TME)GP*CP*GP*GP*A)-3', PROPANE | | Authors: | Dooley, P.D, Zhang, M, Korbel, G.A, Nechev, L.V, Harris, C.M, Stone, M.P, Harris, T.M. | | Deposit date: | 2002-05-22 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Determination of the Conformation of a Trimethylene Interstrand Cross-Link in an Oligodeoxynucleotide Duplex Containing a 5'-d(GpC) Motif
J.AM.CHEM.SOC., 125, 2003
|
|
2ZCY
 
 | | yeast 20S proteasome:syringolin A-complex | | Descriptor: | (2S)-2-[[(2S)-1-[[(5S,8S,9E)-2,7-dioxo-5-propan-2-yl-1,6-diazacyclododeca-3,9-dien-8-yl]amino]-3-methyl-1-oxo-butan-2-yl]carbamoylamino]-3-methyl-butanoic acid, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Dudler, R, Kaiser, M. | | Deposit date: | 2007-11-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A plant pathogen virulence factor inhibits the eukaryotic proteasome by a novel mechanism
Nature, 452, 2008
|
|
3U9E
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoA. | | Descriptor: | ARGININE, CHLORIDE ION, COENZYME A, ... | | Authors: | Tan, K, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-18 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoA.
To be Published
|
|
1I4F
 
 | | CRYSTAL STRUCTURE OF HLA-A*0201/MAGE-A4-PEPTIDE COMPLEX | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Hillig, R.C, Coulie, P.G, Stroobant, V, Saenger, W, Ziegler, A, Huelsmeyer, M. | | Deposit date: | 2001-02-21 | | Release date: | 2001-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structure of HLA-A*0201 in complex with a tumour-specific antigenic peptide encoded by the MAGE-A4 gene.
J.Mol.Biol., 310, 2001
|
|
2A9M
 
 | |
3U18
 
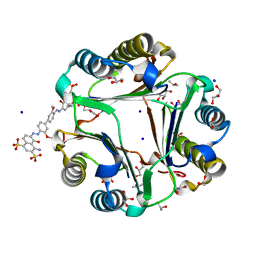 | | Chicago Sky Blue 6B, A Novel Inhibitor for Macrophage Migration Inhibitory Factor | | Descriptor: | 1,2-ETHANEDIOL, 6,6'-[(3,3'-dimethoxybiphenyl-4,4'-diyl)di(E)diazene-2,1-diyl]bis(4-amino-5-hydroxynaphthalene-1,3-disulfonic acid), GLYCEROL, ... | | Authors: | Asojo, O.A. | | Deposit date: | 2011-09-29 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Allosteric Inhibitor of Macrophage Migration Inhibitory Factor (MIF).
J.Biol.Chem., 287, 2012
|
|
2P8S
 
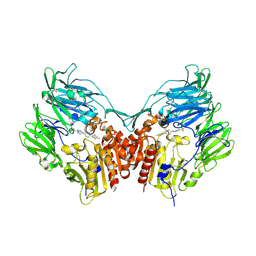 | | Human dipeptidyl peptidase IV/CD26 in complex with a cyclohexalamine inhibitor | | Descriptor: | (1S,2R,5S)-5-[3-(TRIFLUOROMETHYL)-5,6-DIHYDRO[1,2,4]TRIAZOLO[4,3-A]PYRAZIN-7(8H)-YL]-2-(2,4,5-TRIFLUOROPHENYL)CYCLOHEXANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Biftu, T. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational design of a novel, potent, and orally bioavailable cyclohexylamine DPP-4 inhibitor by application of molecular modeling and X-ray crystallography of sitagliptin
Bioorg.Med.Chem.Lett., 17, 2007
|
|
224D
 
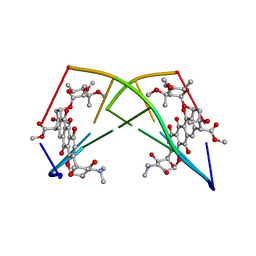 | | DNA-DRUG REFINEMENT: A COMPARISON OF THE PROGRAMS NUCLSQ, PROLSQ, SHELXL93 AND X-PLOR, USING THE LOW TEMPERATURE D(TGATCA)-NOGALAMYCIN STRUCTURE | | Descriptor: | DNA (5'-D(*TP*GP*AP*TP*CP*A)-3'), NOGALAMYCIN | | Authors: | Schuerman, G.S, Smith, C.K, Turkenburg, J.P, Dettmar, A.N, Van Meervelt, L, Moore, M.H. | | Deposit date: | 1995-08-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | DNA-drug refinement: a comparison of the programs NUCLSQ, PROLSQ, SHELXL93 and X-PLOR, using the low-temperature d(TGATCA)-nogalamycin structure.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1DF3
 
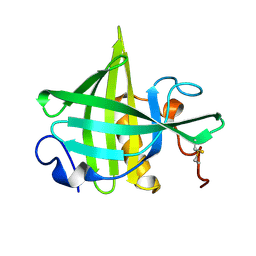 | | SOLUTION STRUCTURE OF A RECOMBINANT MOUSE MAJOR URINARY PROTEIN | | Descriptor: | MAJOR URINARY PROTEIN | | Authors: | Luecke, C, Franzoni, L, Abbate, F, Loehr, F, Ferrari, E, Sorbi, R.T, Rueterjans, H, Spisni, A. | | Deposit date: | 1999-11-17 | | Release date: | 2000-05-10 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a recombinant mouse major urinary protein.
Eur.J.Biochem., 266, 1999
|
|
4EUA
 
 | |
2OYP
 
 | | T Cell Immunoglobulin Mucin-3 Crystal Structure Revealed a Galectin-9-independent Binding Surface | | Descriptor: | Hepatitis A virus cellular receptor 2, SULFATE ION | | Authors: | Cao, E, Ramagopal, U.A, Fedorov, A.A, Fedorov, E.V, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-02-22 | | Release date: | 2007-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | T cell immunoglobulin mucin-3 crystal structure reveals a galectin-9-independent ligand-binding surface
Immunity, 26, 2007
|
|
4EU8
 
 | | Succinyl-CoA:acetate CoA-transferase (AarCH6-S71A) in complex with CoA | | Descriptor: | CHLORIDE ION, COENZYME A, Succinyl-CoA:acetate coenzyme A transferase | | Authors: | Mullins, E.A, Kappock, T.J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.809 Å) | | Cite: | Crystal Structures of Acetobacter aceti Succinyl-Coenzyme A (CoA):Acetate CoA-Transferase Reveal Specificity Determinants and Illustrate the Mechanism Used by Class I CoA-Transferases.
Biochemistry, 51, 2012
|
|
3UBM
 
 | |
1HC8
 
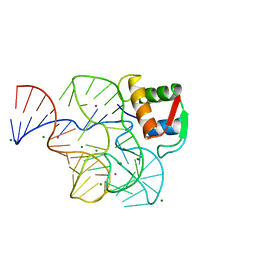 | | CRYSTAL STRUCTURE OF A CONSERVED RIBOSOMAL PROTEIN-RNA COMPLEX | | Descriptor: | 58 NUCLEOTIDE RIBOSOMAL 23S RNA DOMAIN, MAGNESIUM ION, OSMIUM ION, ... | | Authors: | Conn, G.L, Draper, D.E, Lattman, E.E, Gittis, A.G. | | Deposit date: | 2001-04-27 | | Release date: | 2002-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Compact RNA Tertiary Structure Contains a Buried Backbone-K+ Complex
J.Mol.Biol., 318, 2002
|
|
4EUB
 
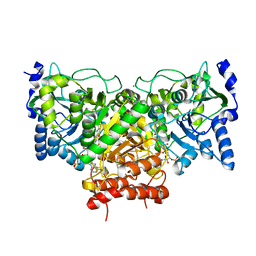 | | Succinyl-CoA:acetate CoA-transferase (AarCH6-E294A) in complex with CoA | | Descriptor: | CHLORIDE ION, COENZYME A, Succinyl-CoA:acetate coenzyme A transferase | | Authors: | Mullins, E.A, Kappock, T.J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Crystal Structures of Acetobacter aceti Succinyl-Coenzyme A (CoA):Acetate CoA-Transferase Reveal Specificity Determinants and Illustrate the Mechanism Used by Class I CoA-Transferases.
Biochemistry, 51, 2012
|
|
4B7F
 
 | | Structure of a liganded bacterial catalase | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Gumiero, A, Walsh, M. | | Deposit date: | 2012-08-20 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Structural and Dynamic Investigation of the Inhibition of Catalase by Nitric Oxide.
Org.Biomol.Chem., 11, 2013
|
|
4EUC
 
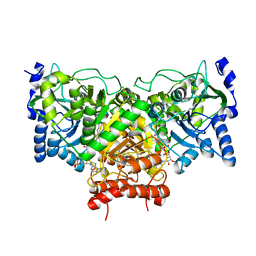 | | Succinyl-CoA:acetate CoA-transferase (AarCH6-E294A) in complex with dethiaacetyl-CoA | | Descriptor: | CHLORIDE ION, Succinyl-CoA:acetate coenzyme A transferase, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(3R)-2,2-dimethyl-3-oxidanyl-4-oxidanylidene-4-[[3-oxidanylidene-3-(4-oxidanylidenepentylamino)propyl]amino]butyl] hydrogen phosphate | | Authors: | Mullins, E.A, Kappock, T.J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Crystal Structures of Acetobacter aceti Succinyl-Coenzyme A (CoA):Acetate CoA-Transferase Reveal Specificity Determinants and Illustrate the Mechanism Used by Class I CoA-Transferases.
Biochemistry, 51, 2012
|
|
4IJH
 
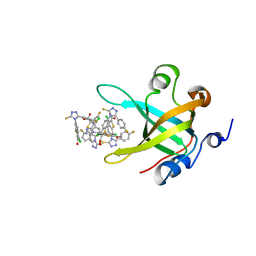 | | Fragment-based Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A | | Descriptor: | 3-chloro-6-[3-(4-fluorophenyl)-5-sulfanyl-4H-1,2,4-triazol-4-yl]-1-benzothiophene-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Patrone, J.D, Kennedy, J.P, Frank, A.O, Vangamudi, B, Pelz, N.F, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2012-12-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A.
ACS MED.CHEM.LETT., 4, 2013
|
|
