4XYY
 
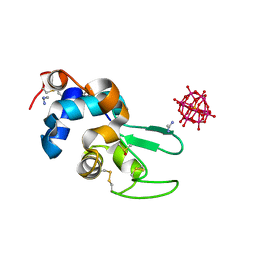 | |
6XWK
 
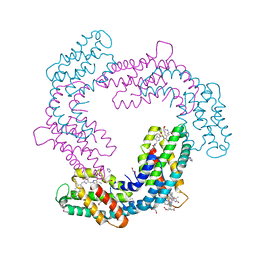 | | Crystal structure of Phormidium rubidum phycocyanin | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Sonani, R.R, Roszak, A.W, Cogdell, R.J, Madamwar, D, Liu, H, Gross, M.L, Blankenship, R.E. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Revisiting high-resolution crystal structure of Phormidium rubidum phycocyanin.
Photosyn. Res., 144, 2020
|
|
4XZW
 
 | | Endo-glucanase chimera C10 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Lee, C.C, Chang, C.J, Ho, T.H.D, Chao, Y.C, Wang, A.H.J. | | Deposit date: | 2015-02-05 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Endo-glucanase chimera C10
To Be Published
|
|
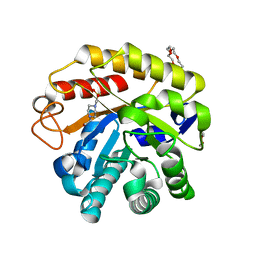
7LOU
 
 | | Crystal structure of Clostridium difficile Toxin B (TcdB) glucosyltransferase in complex with UDP and isofagomine | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, Glucosyltransferase TcdB, ... | | Authors: | Harijan, R.K, Paparella, A.S, Aboulache, B.L, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2021-02-10 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Inhibition of Clostridium difficile TcdA and TcdB toxins with transition state analogues.
Nat Commun, 12, 2021
|
|
8QDB
 
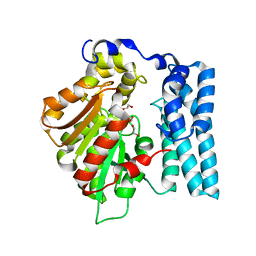 | | Wdyg1p Ser256DHA (PSF) | | Descriptor: | 1,2-ETHANEDIOL, Yellowish-green 1-like protein | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QBI
 
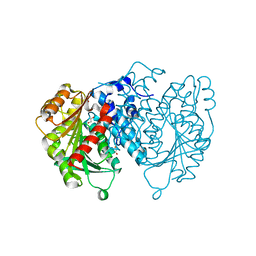 | | AntI in closed state | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Photorhabdus luminescens subsp. laumondii TTO1 complete genome segment 15/17, ... | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8QDA
 
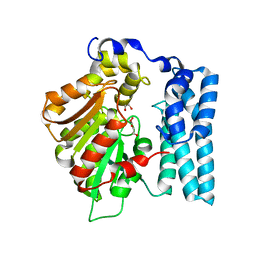 | | Wdyg1p Ser256DHA (PMSF) | | Descriptor: | 1,2-ETHANEDIOL, TRIETHYLENE GLYCOL, Yellowish-green 1-like protein | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
5K6O
 
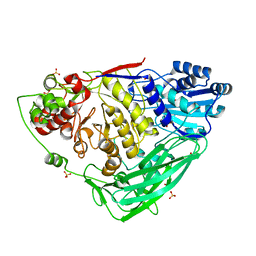 | |
8EQN
 
 | |
1JTJ
 
 | | Solution structure of HIV-1Lai mutated SL1 hairpin | | Descriptor: | HIV-1Lai SL1 | | Authors: | Kieken, F, Arnoult, E, Barbault, F, Paquet, F, Huynh-Dinh, T, Paoletti, J, Genest, D, Lancelot, G. | | Deposit date: | 2001-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HIV-1(Lai) genomic RNA: combined used of NMR and molecular dynamics simulation for studying
the structure and internal dynamics of a mutated SL1 hairpin.
EUR.BIOPHYS.J., 31, 2002
|
|
403D
 
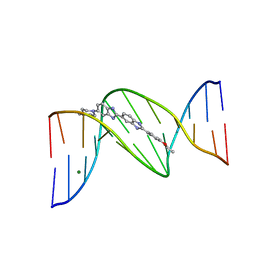 | | 5'-D(*CP*GP*CP*(HYD)AP*AP*AP*TP*TP*TP*GP*CP*G)-3', 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(IGU)P*AP*AP*TP*TP*TP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Robinson, H, Gao, Y.-G, Bauer, C, Roberts, C, Switzer, C, Wang, A.H.-J. | | Deposit date: | 1998-06-10 | | Release date: | 1998-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 2'-Deoxyisoguanosine adopts more than one tautomer to form base pairs with thymidine observed by high-resolution crystal structure analysis.
Biochemistry, 37, 1998
|
|
9RSA
 
 | |
6NM7
 
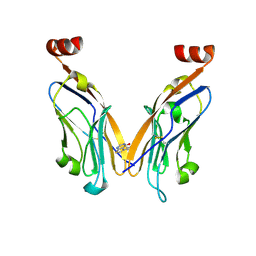 | | PD-L1 IgV domain bound to fragment | | Descriptor: | 5-phenylthieno[2,3-d]pyrimidin-4(3H)-one, Programmed cell death 1 ligand 1 | | Authors: | Perry, E, Zhao, B. | | Deposit date: | 2019-01-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.426 Å) | | Cite: | Fragment-based screening of programmed death ligand 1 (PD-L1).
Bioorg. Med. Chem. Lett., 29, 2019
|
|
9RYG
 
 | |
9MG6
 
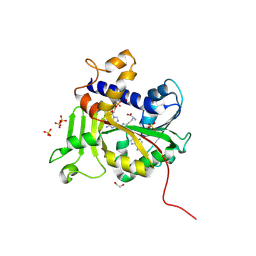 | | Structure of Saccharomyces cerevisiae mRNA cap (guanine-N7) methyltransferase variant, Abd1-K163R-K311R-F387Y-Y416F, in complex with sinefungin and GTP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-TRIPHOSPHATE, SINEFUNGIN, ... | | Authors: | Nilson, D.J, Ghosh, A. | | Deposit date: | 2024-12-10 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for sensitivity and acquired resistance of fungal cap guanine-N7 methyltransferases to the antifungal antibiotic sinefungin.
Nucleic Acids Res., 53, 2025
|
|
9QYT
 
 | | Crystal structure of leaf branch compost cutinase variant ICCG L50Y T110E | | Descriptor: | 1,2-ETHANEDIOL, Leaf-branch compost cutinase, SODIUM ION | | Authors: | Bischoff, D, Walla, B, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2025-04-20 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Application of a Rational Crystal Contact Engineering Strategy on a Poly(ethylene terephthalate)-Degrading Cutinase.
Bioengineering (Basel), 12, 2025
|
|
9MFP
 
 | | Cat DHX9 in Complex with Compound 1 and ADP | | Descriptor: | 1,2-ETHANEDIOL, 1-ethyl-N-[3-(methanesulfonamido)phenyl]-5-methyl-1H-pyrazole-3-carboxamide, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lockbaum, G.J, Lee, Y.-T, Sickmier, E.A, Boriack-Sjodin, P.A, Grigoriu, S. | | Deposit date: | 2024-12-10 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery of ATX968: An Orally Available Allosteric Inhibitor of DHX9.
J.Med.Chem., 68, 2025
|
|
8ERE
 
 | |
9NAW
 
 | |
9VCK
 
 | | Cryo-EM structure of SARS-CoV-2 nsp10/nsp14:RNA:SMP complex | | Descriptor: | CALCIUM ION, Guanine-N7 methyltransferase nsp14, Non-structural protein 10, ... | | Authors: | Wang, J, Lou, Z, Liu, D. | | Deposit date: | 2025-06-06 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Structural Basis and Rational Design of Nucleotide Analogue Inhibitor Evading the SARS-CoV-2 Proofreading Enzyme.
J.Am.Chem.Soc., 147, 2025
|
|
9V7W
 
 | |
9VCL
 
 | | Cryo-EM structure of SARS-CoV-2 nsp10/nsp14:RNA:ATMP complex | | Descriptor: | CALCIUM ION, Guanine-N7 methyltransferase nsp14, Non-structural protein 10, ... | | Authors: | Wang, J, Lou, Z, Liu, D. | | Deposit date: | 2025-06-06 | | Release date: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural Basis and Rational Design of Nucleotide Analogue Inhibitor Evading the SARS-CoV-2 Proofreading Enzyme.
J.Am.Chem.Soc., 147, 2025
|
|
9UJ2
 
 | | 14-3-3 zeta chimera with the S202R peptide of SARS-CoV-2 N (residues 200-213) | | Descriptor: | 1,2-ETHANEDIOL, 14-3-3 protein zeta/delta,Peptide from Nucleoprotein, DI(HYDROXYETHYL)ETHER | | Authors: | Boyko, K.M, Matyuta, I.O, Minyaev, M.E, Perfilova, K.V, Sluchanko, N.N. | | Deposit date: | 2025-04-16 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structure reveals enhanced 14-3-3 binding by a mutant SARS-CoV-2 nucleoprotein variant with improved replicative fitness.
Biochem.Biophys.Res.Commun., 767, 2025
|
|
9QYU
 
 | | Crystal structure of leaf branch compost cutinase quintuple variant ICCG L50Y | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Leaf-branch compost cutinase | | Authors: | Bischoff, D, Walla, B, Dietrich, A.-M, Janowski, R, Niessing, D, Weuster-Botz, D. | | Deposit date: | 2025-04-21 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Application of a Rational Crystal Contact Engineering Strategy on a Poly(ethylene terephthalate)-Degrading Cutinase.
Bioengineering (Basel), 12, 2025
|
|
9UET
 
 | | Cryo-EM structure of human choline-phosphotransferase 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Cholinephosphotransferase 1, MAGNESIUM ION | | Authors: | He, Y.L, Qian, H.W. | | Deposit date: | 2025-04-09 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM structure of human choline-phosphotransferase 1
To Be Published
|
|
