5BR4
 
 | | E. coli lactaldehyde reductase (FucO) M185C mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Lactaldehyde reductase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Mutations in adenine-binding pockets enhance catalytic properties of NAD(P)H-dependent enzymes.
Protein Eng.Des.Sel., 29, 2016
|
|
8FSG
 
 | |
8FSM
 
 | |
8FSN
 
 | |
5BRT
 
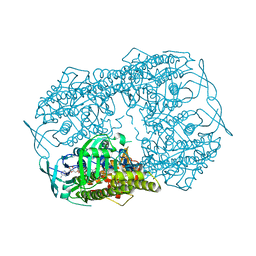 | | Crystal Structure of 2-hydroxybiphenyl 3-monooxygenase from Pseudomonas azelaica with 2-hydroxybiphenyl in the active site | | Descriptor: | 2-HYDROXYBIPHENYL, 2-hydroxybiphenyl-3-monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Kanteev, M, Bregman-Cohen, A, Deri, B, Adir, N, Fishman, A. | | Deposit date: | 2015-06-01 | | Release date: | 2015-08-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A crystal structure of 2-hydroxybiphenyl 3-monooxygenase with bound substrate provides insights into the enzymatic mechanism.
Biochim.Biophys.Acta, 1854, 2015
|
|
6UW4
 
 | | Cryo-EM structure of human TRPV3 determined in lipid nanodisc | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily V member 3, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, Yuan, P. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Gating of human TRPV3 in a lipid bilayer.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8FSW
 
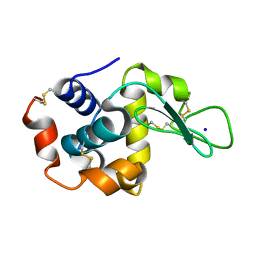 | |
8FSU
 
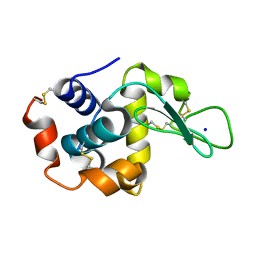 | |
8FSV
 
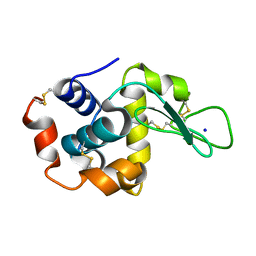 | |
8FST
 
 | |
8FT0
 
 | |
8FSY
 
 | |
5BTV
 
 | | Crystal structure of human 14-3-3 sigma in complex with a Tau-protein peptide surrounding pS324 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ottmann, C, Schumacher, B, Bartel, M. | | Deposit date: | 2015-06-03 | | Release date: | 2016-07-20 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Involvement of 14-3-3 in tubulin instability and impaired axon development is mediated by Tau.
Faseb J., 29, 2015
|
|
8FSX
 
 | |
8FT3
 
 | |
8FT1
 
 | |
4Q0I
 
 | | Deinococcus radiodurans BphP PAS-GAF D207A mutant | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium-2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome, ISOPROPYL ALCOHOL | | Authors: | Burgie, E.S, Vierstra, R.D. | | Deposit date: | 2014-04-02 | | Release date: | 2014-07-16 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (1.744 Å) | | Cite: | Crystallographic and Electron Microscopic Analyses of a Bacterial Phytochrome Reveal Local and Global Rearrangements during Photoconversion.
J.Biol.Chem., 289, 2014
|
|
8FZ8
 
 | | Structure of cytochrome P450sky2 | | Descriptor: | Cytochrome P450, OCTANOIC ACID (CAPRYLIC ACID), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murarka, V.C, Poulos, T.L. | | Deposit date: | 2023-01-27 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Biosynthesis of a new skyllamycin in Streptomyces nodosus : a cytochrome P450 forms an epoxide in the cinnamoyl chain.
Org.Biomol.Chem., 22, 2024
|
|
6UQZ
 
 | |
6V13
 
 | | immune receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen beta 74cit69-81, G08 TCR alpha chain, ... | | Authors: | Lim, J.J, Rossjohn, J. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The shared susceptibility epitope of HLA-DR4 binds citrullinated self-antigens and the TCR.
Sci Immunol, 6, 2021
|
|
5APE
 
 | | Hen Egg White Lysozyme reference dataset odd frames | | Descriptor: | LYSOZYME C, SODIUM ION | | Authors: | Lundholm, I, Rodilla, H, Wahlgren, W.Y, Duelli, A, Bourenkov, G, Vukusic, J, Friedman, R, Stake, J, Schneider, T, Katona, G. | | Deposit date: | 2015-09-15 | | Release date: | 2016-01-13 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Terahertz Radiation Induces Non-Thermal Structural Changes Associated with Frohlich Condensation in a Protein Crystal
Struct.Dyn., 2, 2015
|
|
4Q1U
 
 | | Serum paraoxonase-1 by directed evolution with the K192Q mutation | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ben-David, M, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2014-04-04 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Catalytic stimulation by restrained active-site floppiness-the case of high density lipoprotein-bound serum paraoxonase-1.
J.Mol.Biol., 427, 2015
|
|
5B66
 
 | | Crystal structure analysis of Photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Tanaka, A, Fukushima, Y, Kamiya, N. | | Deposit date: | 2016-05-25 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Two Different Structures of the Oxygen-Evolving Complex in the Same Polypeptide Frameworks of Photosystem II
J. Am. Chem. Soc., 139, 2017
|
|
6US4
 
 | | MTH1 in complex with compound 32 | | Descriptor: | 5-(2,3-dichlorophenyl)[1,2,4]triazolo[1,5-a]pyridin-2-amine, 7,8-dihydro-8-oxoguanine triphosphatase | | Authors: | Newby, Z.E.R, Lansdon, E.B. | | Deposit date: | 2019-10-24 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95032907 Å) | | Cite: | Discovery of Potent and Selective MTH1 Inhibitors for Oncology: Enabling Rapid Target (In)Validation.
Acs Med.Chem.Lett., 11, 2020
|
|
6V2N
 
 | | Crystal structure of E. coli phosphoenolpyruvate carboxykinase mutant Lys254Ser | | Descriptor: | ACETATE ION, CALCIUM ION, Phosphoenolpyruvate carboxykinase (ATP) | | Authors: | Sokaribo, A.S, Cotelesage, J.H, Novakovski, B, Goldie, H, Sanders, D. | | Deposit date: | 2019-11-25 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Kinetic and structural analysis of Escherichia coli phosphoenolpyruvate carboxykinase mutants.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
