6EP5
 
 | |
7ONC
 
 | | Crystal structure of the computationally designed SAKe6BE protein | | Descriptor: | SAKe6BE | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
5TDO
 
 | |
7JY1
 
 | | Structure of HbA with compound 19 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Jasti, J. | | Deposit date: | 2020-08-28 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | PF-07059013: A Noncovalent Modulator of Hemoglobin for Treatment of Sickle Cell Disease.
J.Med.Chem., 64, 2021
|
|
7ONH
 
 | | Crystal structure of the computationally designed SAKe6BE-L3 protein | | Descriptor: | SAKe6BE-L3, SULFATE ION | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7YPZ
 
 | | Zafirlukast in complex with CRM1-Ran-RanBP1 | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2022-08-05 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Searching for Novel Noncovalent Nuclear Export Inhibitors through a Drug Repurposing Approach.
J.Med.Chem., 66, 2023
|
|
7OL2
 
 | | Crystal structure of mouse contactin 1 immunoglobulin domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Contactin-1, ... | | Authors: | Chataigner, L.M.P, Janssen, B.J.C. | | Deposit date: | 2021-05-19 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structural insights into the contactin 1 - neurofascin 155 adhesion complex.
Nat Commun, 13, 2022
|
|
8PGB
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2850 | | Descriptor: | 3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-7-[(1~{S})-1-(5-methyl-1,2,4-oxadiazol-3-yl)ethyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2850
To Be Published
|
|
5TE0
 
 | | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with BIBF 1120 | | Descriptor: | AP2-associated protein kinase 1, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Counago, R.M, Elkins, J.M, Bountra, C, Arruda, P, Edwards, A.M, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-20 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with BIBF 1120
To Be Published
|
|
7ON6
 
 | | Crystal structure of the computationally designed SAKe6AE protein | | Descriptor: | SAKe6AE, SULFATE ION | | Authors: | Wouters, S.M.L, Noguchi, H, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
6EK9
 
 | | Cytosolic copper storage protein Csp from Streptomyces lividans: Cu loaded form | | Descriptor: | COPPER (I) ION, Cytosolic copper storage protein | | Authors: | Straw, M.L, Chaplin, A.K, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-09-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A cytosolic copper storage protein provides a second level of copper tolerance in Streptomyces lividans.
Metallomics, 10, 2018
|
|
8PGU
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3394 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(6-oxidanylidene-1~{H}-pyridin-3-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3394
To Be Published
|
|
5EGL
 
 | | The structural and biochemical characterization of acyl-coa hydrolase from Staphylococcus aureus in complex with Butyryl Coenzyme A, Coenzyme A, and Coenzyme A disulfide | | Descriptor: | Acyl CoA Hydrolase, Butyryl Coenzyme A, COENZYME A, ... | | Authors: | Khandokar, Y.B, Srivastava, P.S, Forwood, J.K. | | Deposit date: | 2015-10-27 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural and biochemical characterization of acyl-coa hydrolase from Staphylococcus aureus
To Be Published
|
|
8PNO
 
 | |
5TEE
 
 | | Crystal structure of Gemin5 WD40 repeats in apo form | | Descriptor: | GLYCEROL, Gem-associated protein 5, SODIUM ION, ... | | Authors: | Chao, X, Tempel, W, Bian, C, Cerovina, T, He, H, Walker, J.R, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-09-21 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into Gemin5-guided selection of pre-snRNAs for snRNP assembly.
Genes Dev., 30, 2016
|
|
7YSZ
 
 | | Spiroplasma melliferum FtsZ bound to GDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-08-13 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3000412 Å) | | Cite: | Structural basis of kinetic polarity of FtsZ from a cell wall less bacterium Spiroplasma
To Be Published
|
|
7OPV
 
 | | Crystal structure of the computationally designed SAKe6BE-3HH protein, alternative packing | | Descriptor: | SAKe6BE-3HH | | Authors: | Wouters, S.M.L, Noguchi, H, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-06-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7YOP
 
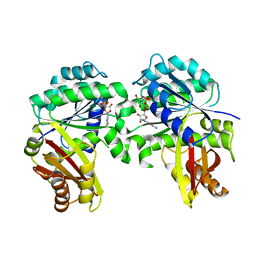 | | Spiroplasma melliferum FtsZ bound to GMPPNP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chakraborty, J, Pananghat, G. | | Deposit date: | 2022-08-01 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.20003128 Å) | | Cite: | Structural basis of kinetic polarity of FtsZ from a cell wall-less bacterium Spiroplasma
To Be Published
|
|
7OP4
 
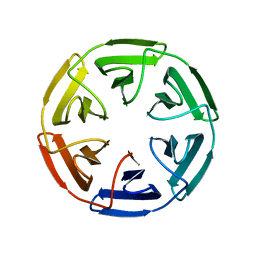 | |
8PGX
 
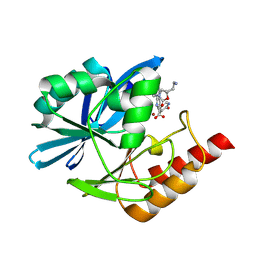 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3461 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(5-fluoranyl-6-oxidanylidene-1~{H}-pyridin-3-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3461
To Be Published
|
|
6XTM
 
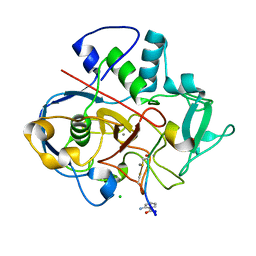 | |
7OWP
 
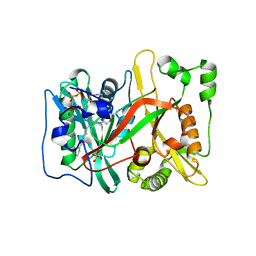 | | HsNMT1 in complex with both MyrCoA and ACE-G-(L-ORN)SFSKPR | | Descriptor: | ACE-GLY-ORN-SER-PHE-SER-LYS-PRO-ARG, GLYCEROL, Glycylpeptide N-tetradecanoyltransferase 1, ... | | Authors: | Dian, C, Giglione, C, Meinnel, T. | | Deposit date: | 2021-06-18 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural and Large-scale Analysis Unveil the Intertwined Paths Promoting NMT-catalyzed Lysine and Glycine Myristoylation.
J.Mol.Biol., 434, 2022
|
|
8PIP
 
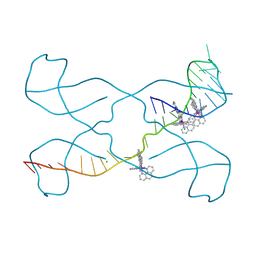 | |
5ERD
 
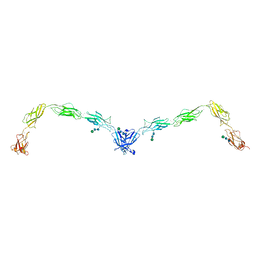 | | Crystal structure of human Desmoglein-2 ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of adhesive binding by desmocollins and desmogleins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7OVU
 
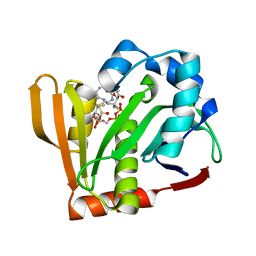 | |
