3TRZ
 
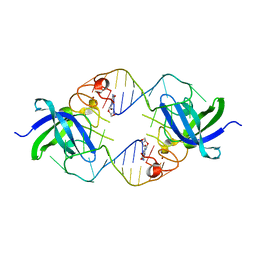 | | Mouse Lin28A in complex with let-7d microRNA pre-element | | Descriptor: | Protein lin-28 homolog A, RNA (5'-R(*GP*GP*GP*CP*AP*GP*GP*GP*AP*UP*UP*UP*UP*GP*CP*CP*CP*GP*GP*AP*G)-3'), ZINC ION | | Authors: | Nam, Y, Sliz, P. | | Deposit date: | 2011-09-11 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for Interaction of let-7 MicroRNAs with Lin28.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TSV
 
 | |
3U2K
 
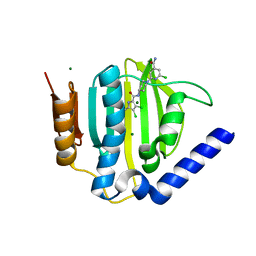 | | S. aureus GyrB ATPase domain in complex with a small molecule inhibitor | | Descriptor: | 2-chloro-6-(4-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}piperidin-1-yl)pyridine-4-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
3TYQ
 
 | |
3TYI
 
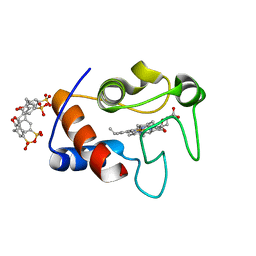 | | Crystal Structure of Cytochrome c - p-Sulfonatocalix[4]arene Complexes | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, Cytochrome c iso-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mc Govern, R.E, Fernandes, H, Khan, A.R, Crowley, P.B. | | Deposit date: | 2011-09-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Protein camouflage in cytochrome c-calixarene complexes.
Nat Chem, 4, 2012
|
|
3QOR
 
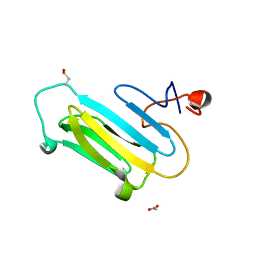 | |
3Q68
 
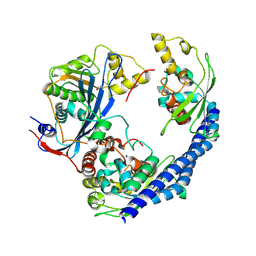 | |
3Q66
 
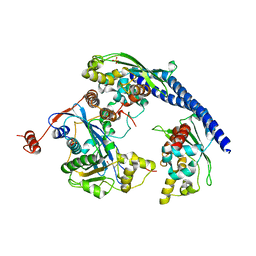 | |
3RRZ
 
 | | H-Ras in 70% glycerol: one of 10 in MSCS set | | Descriptor: | CALCIUM ION, GLYCEROL, GTPase HRas, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
3RS5
 
 | | H-Ras soaked in 55% dimethylformamide: 1 of 10 in MSCS set | | Descriptor: | CALCIUM ION, DIMETHYLFORMAMIDE, GTPase HRas, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
3RCP
 
 | | Crystal structure of the FAPP1 pleckstrin homology domain | | Descriptor: | GLYCEROL, Pleckstrin homology domain-containing family A member 3 | | Authors: | Roy, S, He, J, Kutateladze, T.G. | | Deposit date: | 2011-03-31 | | Release date: | 2011-04-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis of Phosphatidylinositol 4-Phosphate and ARF1 GTPase Recognition by the FAPP1 Pleckstrin Homology (PH) Domain.
J.Biol.Chem., 286, 2011
|
|
3R36
 
 | | Crystal structure of Arthrobacter sp. strain SU 4-hydroxybenzoyl CoA thioesterase mutant E73Q complexed with 4-hydroxybenzoic acid | | Descriptor: | 4-hydroxybenzoyl-CoA thioesterase, P-HYDROXYBENZOIC ACID | | Authors: | Holden, H.M, Thoden, J.B, Song, F, Zhuang, Z, Trujillo, M, Dunaway-Mariano, D. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Catalytic Mechanism of the Hotdog-fold Enzyme Superfamily 4-Hydroxybenzoyl-CoA Thioesterase from Arthrobacter sp. Strain SU.
Biochemistry, 51, 2012
|
|
3RH1
 
 | | X-ray Structure of a cis-proline (P114) to alanine variant of Ribonuclease A | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic | | Authors: | Merlino, A, Balsamo, A, Mazzarella, L, Sica, F. | | Deposit date: | 2011-04-11 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chain termini cross-talk in the swapping process of bovine pancreatic ribonuclease.
Biochimie, 94, 2012
|
|
3RM4
 
 | | AMCase in complex with Compound 1 | | Descriptor: | 5-{4-[2-(4-bromophenoxy)ethyl]piperazin-1-yl}-4H-1,2,4-triazol-3-amine, Acidic mammalian chitinase | | Authors: | Olland, A. | | Deposit date: | 2011-04-20 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and Characterization of Acidic Mammalian Chitinase Inhibitors
J.Med.Chem., 53, 2010
|
|
3RRY
 
 | | H-Ras crosslinked control, soaked in aqueous solution: one of 10 in MSCS set | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
3RS4
 
 | | H-Ras soaked in 60% 1,6-hexanediol: 1 of 10 in MSCS set | | Descriptor: | CALCIUM ION, GTPase HRas, HEXANE-1,6-DIOL, ... | | Authors: | Mattos, C, Buhrman, G, Kearney, B. | | Deposit date: | 2011-05-02 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of Binding Site Hot Spots on the Surface of Ras GTPase.
J.Mol.Biol., 413, 2011
|
|
3U8X
 
 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-29) of drosophila Ciboulot and the C-terminal domain (residues 18-44) of bovine Thymosin-beta4, bound to G-actin-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
3TS0
 
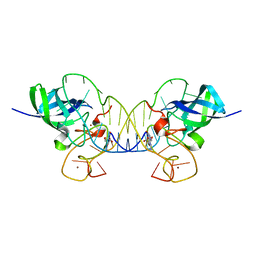 | | Mouse Lin28A in complex with let-7f-1 microRNA pre-element | | Descriptor: | Protein lin-28 homolog A, RNA (5'-R(*GP*GP*GP*GP*UP*AP*GP*UP*GP*AP*UP*UP*UP*UP*AP*CP*CP*CP*UP*GP*GP*AP*G)-3'), ZINC ION | | Authors: | Nam, Y, Sliz, P. | | Deposit date: | 2011-09-11 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.763 Å) | | Cite: | Molecular Basis for Interaction of let-7 MicroRNAs with Lin28.
Cell(Cambridge,Mass.), 147, 2011
|
|
3TVL
 
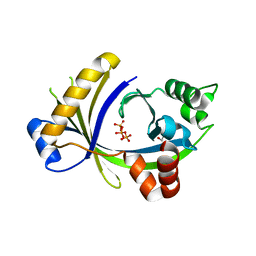 | | Complex between the human thiamine triphosphatase and triphosphate | | Descriptor: | 1,2-ETHANEDIOL, TRIPHOSPHATE, Thiamine-triphosphatase | | Authors: | Delvaux, D, Herman, R, Sauvage, E, Wins, P, Bettendorff, L, Charlier, P, Kerff, F. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of specificity and catalytic mechanism in mammalian 25-kDa thiamine triphosphatase.
Biochim.Biophys.Acta, 1830, 2013
|
|
3TYO
 
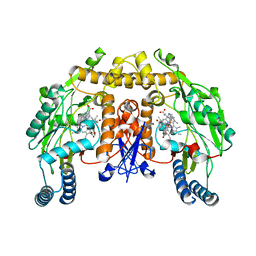 | | Structure of neuronal nitric oxide synthase heme domain in complex with 6-(((3S,4S)-4-(2-((furan-2-ylmethyl)amino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4S)-4-{2-[(furan-2-ylmethyl)amino]ethoxy}pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2011-09-26 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Intramolecular hydrogen bonding: A potential strategy for more bioavailable inhibitors of neuronal nitric oxide synthase.
Bioorg.Med.Chem., 20, 2012
|
|
3TYM
 
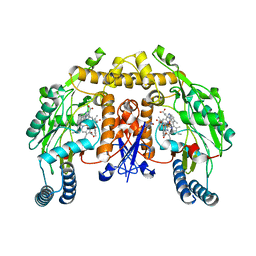 | | Structure of neuronal nitric oxide synthase heme domain in complex with 6-(((3S,4S)-4-(2-((2-methoxybenzyl)amino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4S)-4-{2-[(2-methoxybenzyl)amino]ethoxy}pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2011-09-26 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intramolecular hydrogen bonding: A potential strategy for more bioavailable inhibitors of neuronal nitric oxide synthase.
Bioorg.Med.Chem., 20, 2012
|
|
3TWY
 
 | | RAT PKC C2 DOMAIN BOUND TO PB | | Descriptor: | LEAD (II) ION, Protein kinase C alpha type, SULFATE ION | | Authors: | Li, P. | | Deposit date: | 2011-09-22 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pb2+ as Modulator of Protein-Membrane Interactions.
J.Am.Chem.Soc., 133, 2011
|
|
3L5C
 
 | | Structure of BACE Bound to SCH723871 | | Descriptor: | 1-(4-cyanophenyl)-3-(4-{[(2Z,4R)-2-imino-4-methyl-4-(2-methylpropyl)-5-oxoimidazolidin-1-yl]methyl}benzyl)urea, Beta-secretase 1, D(-)-TARTARIC ACID | | Authors: | Strickland, C, Zhu, Z. | | Deposit date: | 2009-12-21 | | Release date: | 2010-02-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
J.Med.Chem., 53, 2010
|
|
5HF3
 
 | | Crystal structure of C-terminal modified Tau peptide-hybrid 201D with 14-3-3sigma | | Descriptor: | 14-3-3 protein sigma, modified Tau peptide | | Authors: | Bartel, M, Milroy, L.G, Brunsveld, L, Ottmann, C. | | Deposit date: | 2016-01-06 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
3LFV
 
 | | crystal structure of unliganded PDE5A GAF domain | | Descriptor: | cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2010-01-18 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.
J.Biol.Chem., 285, 2010
|
|
