8AGU
 
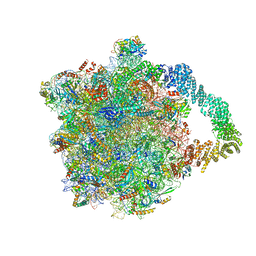 | | Yeast RQC complex in state E | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Tesina, P, Buschauer, R, Beckmann, R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of eIF5A-dependent CAT tailing in eukaryotic ribosome-associated quality control.
Mol.Cell, 83, 2023
|
|
6LHV
 
 | | Structure of FANCA and FANCG Complex | | Descriptor: | Fanconi anemia complementation group A, Fanconi anemia complementation group G | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.59 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
7B6A
 
 | | BK Polyomavirus VP1 pentamer core (residues 30-299) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, IODIDE ION, ... | | Authors: | Osipov, E.M, Munawar, A, Beelen, S, Strelkov, S.V. | | Deposit date: | 2020-12-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Discovery of novel druggable pockets on polyomavirus VP1 through crystallographic fragment-based screening to develop capsid assembly inhibitors.
Rsc Chem Biol, 3, 2022
|
|
7B6C
 
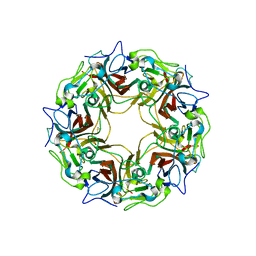 | | BK Polyomavirus VP1 pentamer fusion with long C-terminal extended arm | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, Major capsid protein VP1,Major capsid protein VP1 | | Authors: | Osipov, E.M, Beelen, S, Strelkov, S.V. | | Deposit date: | 2020-12-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Discovery of novel druggable pockets on polyomavirus VP1 through crystallographic fragment-based screening to develop capsid assembly inhibitors.
Rsc Chem Biol, 3, 2022
|
|
4BL7
 
 | |
4L39
 
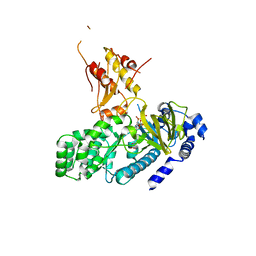 | | Crystal structure of GH3.12 from Arabidopsis thaliana in complex with AMPCPP and salicylate | | Descriptor: | 2-HYDROXYBENZOIC ACID, 4-substituted benzoates-glutamate ligase GH3.12, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Zubieta, C, Jez, J.M, Brown, E, Marcellin, R, Kapp, U, Round, A, Westfall, C. | | Deposit date: | 2013-06-05 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Determination of the GH3.12 protein conformation through HPLC-integrated SAXS measurements combined with X-ray crystallography.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4QFT
 
 | |
4D10
 
 | | Crystal structure of the COP9 signalosome | | Descriptor: | COP9 SIGNALOSOME COMPLEX SUBUNIT 1, COP9 SIGNALOSOME COMPLEX SUBUNIT 2, COP9 SIGNALOSOME COMPLEX SUBUNIT 3, ... | | Authors: | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal Structure of the Human Cop9 Signalosome
Nature, 512, 2014
|
|
5CLQ
 
 | | Ran Y39A in complex with GPPNHP and RanBD1 | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Vetter, I.R, Brucker, S. | | Deposit date: | 2015-07-16 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Catalysis of GTP Hydrolysis by Small GTPases at Atomic Detail by Integration of X-ray Crystallography, Experimental, and Theoretical IR Spectroscopy.
J.Biol.Chem., 290, 2015
|
|
3BWQ
 
 | | Structure of free SV40 VP1 pentamer | | Descriptor: | Capsid protein VP1 | | Authors: | Neu, U, Stehle, T. | | Deposit date: | 2008-01-10 | | Release date: | 2008-03-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of GM1 ganglioside recognition by simian virus 40.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
7W75
 
 | |
7W76
 
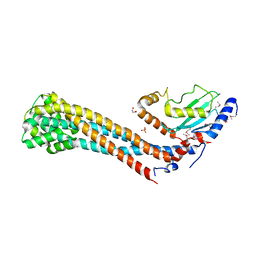 | | Crystal structure of the K. lactis Bre1 RBD in complex with Rad6, crystal form II | | Descriptor: | E3 ubiquitin-protein ligase BRE1, GLYCEROL, SULFATE ION, ... | | Authors: | Shi, M, Zhao, J, Xiang, S. | | Deposit date: | 2021-12-03 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis for the Rad6 activation by the Bre1 N-terminal domain.
Elife, 12, 2023
|
|
3I8Z
 
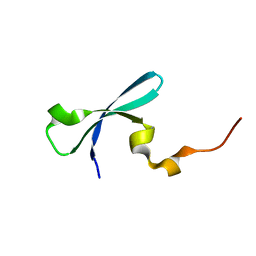 | | Crystal structure of human chromobox homolog 4 (CBX4) | | Descriptor: | E3 SUMO-protein ligase CBX4 | | Authors: | Amaya, M.F, Zhihong, L, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-10 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of human chromobox homolog 4 (CBX4)
To be Published
|
|
3BWR
 
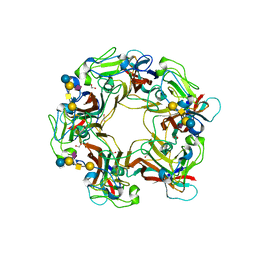 | | SV40 VP1 pentamer in complex with GM1 oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Neu, U, Stehle, T. | | Deposit date: | 2008-01-10 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of GM1 ganglioside recognition by simian virus 40.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
8WMS
 
 | |
6XQJ
 
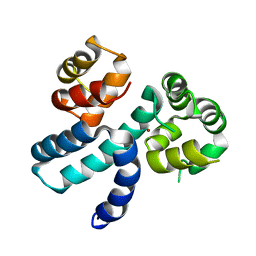 | | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A | | Descriptor: | Protein Vpr,UV excision repair protein RAD23 homolog A, ZINC ION | | Authors: | Byeon, I.-J.L, Calero, G, Wu, Y, Byeon, C.H, Gronenborn, A.M. | | Deposit date: | 2020-07-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.
Nat Commun, 12, 2021
|
|
3I2N
 
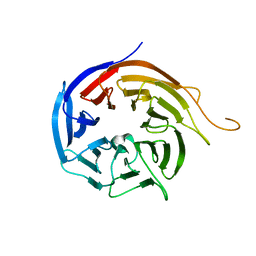 | | Crystal Structure of WD40 repeats protein WDR92 | | Descriptor: | WD repeat-containing protein 92 | | Authors: | Amaya, M.F, Li, Z, He, H, Seitova, A, Ni, S, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
7ZHJ
 
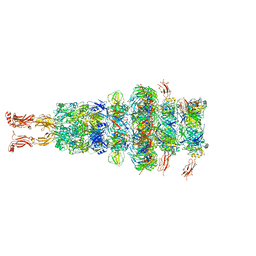 | | Tail tip of siphophage T5 : tip proteins | | Descriptor: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-06 | | Release date: | 2023-02-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7ZQB
 
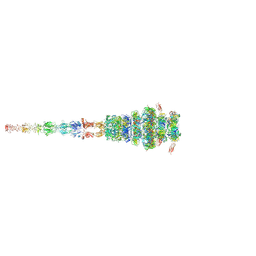 | | Tail tip of siphophage T5 : full structure | | Descriptor: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7ZQP
 
 | | Tail tip of siphophage T5 : open cone after interaction with bacterial receptor FhuA | | Descriptor: | Probable baseplate hub protein, Probable tape measure protein | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
7ZN2
 
 | | Tail tip of siphophage T5 : full complex after interaction with its bacterial receptor FhuA | | Descriptor: | Distal tail protein, L-shaped tail fiber protein p132, Minor tail protein, ... | | Authors: | Linares, R, Arnaud, C.A, Effantin, G, Darnault, C, Epalle, N, Boeri Erba, E, Schoehn, G, Breyton, C. | | Deposit date: | 2022-04-20 | | Release date: | 2023-02-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Structural basis of bacteriophage T5 infection trigger and E. coli cell wall perforation.
Sci Adv, 9, 2023
|
|
5A5H
 
 | |
2J5B
 
 | | Structure of the Tyrosyl tRNA synthetase from Acanthamoeba polyphaga Mimivirus complexed with tyrosynol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, TYROSYL-TRNA SYNTHETASE | | Authors: | Abergel, C, Rudinger-thirion, J, Giege, R, Claverie, J.M. | | Deposit date: | 2006-09-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Virus-Encoded Aminoacyl-tRNA Synthetases: Structural and Functional Characterization of Mimivirus Tyrrs and Metrs.
J.Virol., 81, 2007
|
|
7R9W
 
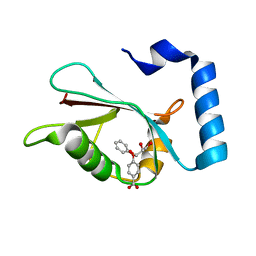 | | LC3A in complex with Fragment 1-1 | | Descriptor: | 4-phenoxybenzoic acid, GLYCEROL, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
7R9Z
 
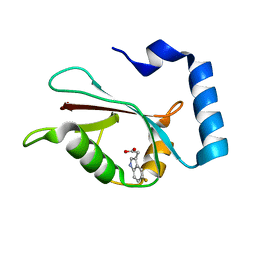 | | LC3A in complex with Fragment 2-3 | | Descriptor: | (5-fluoro-1H-indol-3-yl)acetic acid, Microtubule-associated proteins 1A/1B light chain 3A | | Authors: | Rouge, L, Steffek, M, Helgason, E, Dueber, E, Mulvihill, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A Multifaceted Hit-Finding Approach Reveals Novel LC3 Family Ligands.
Biochemistry, 62, 2023
|
|
