2FUK
 
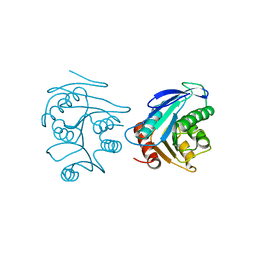 | | Crystal structure of XC6422 from Xanthomonas campestris: a member of a/b serine hydrolase without lid at 1.6 resolution | | Descriptor: | XC6422 protein | | Authors: | Yang, C.Y, Chin, K.H, Chou, C.C, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2006-01-27 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of XC6422 from Xanthomonas campestris at 1.6 A resolution: a small serine alpha/beta-hydrolase
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3ZD4
 
 | |
2FKX
 
 | |
2F8B
 
 | |
4C2K
 
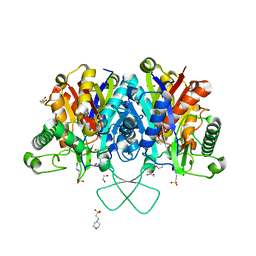 | | Crystal structure of human mitochondrial 3-ketoacyl-CoA thiolase | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kiema, T.-R, Harijan, R.K, Wierenga, R.K. | | Deposit date: | 2013-08-19 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Human Mitochondrial 3-Ketoacyl-Coa Thiolase (T1): Insight Into the Reaction Mechanism of its Thiolase and Thioesterase Activities
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2Z27
 
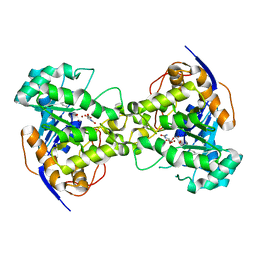 | | Thr109Ser dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
4A3U
 
 | |
3U1R
 
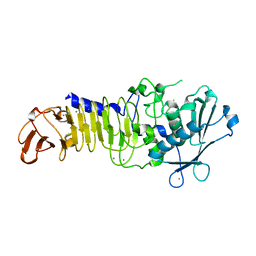 | | Structure Analysis of A New Psychrophilic Marine Protease | | Descriptor: | Alkaline metalloprotease, CALCIUM ION, ZINC ION | | Authors: | Zhang, S.-C, Sun, M, Tang, L, Wang, Q.-H, Hao, J.-H, Han, Y, Hu, X.-J, Zhou, M, Lin, S.-X. | | Deposit date: | 2011-09-30 | | Release date: | 2012-10-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure analysis of a new psychrophilic marine protease.
Plos One, 6, 2011
|
|
3MYN
 
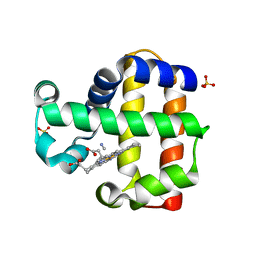 | | Mutation of Methionine-86 in Dehaloperoxidase-hemoglobin: Effects of the Asp-His-Fe Triad in a 3/3 Globin | | Descriptor: | CYANIDE ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bowden, E.F, de Serrano, V.S, D'Antonio, E.L, Franzen, S. | | Deposit date: | 2010-05-10 | | Release date: | 2011-04-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Functional consequences of the creation of an Asp-His-Fe triad in a 3/3 globin.
Biochemistry, 50, 2011
|
|
3MYM
 
 | | Mutation of Methionine-86 in Dehaloperoxidase-hemoglobin: Effects of the Asp-His-Fe Triad in a 3/3 Globin | | Descriptor: | CYANIDE ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | de Serrano, V.S, D'Antonio, E.L, Franzen, S, Bowden, E.F. | | Deposit date: | 2010-05-10 | | Release date: | 2011-04-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Functional consequences of the creation of an Asp-His-Fe triad in a 3/3 globin.
Biochemistry, 50, 2011
|
|
3PRC
 
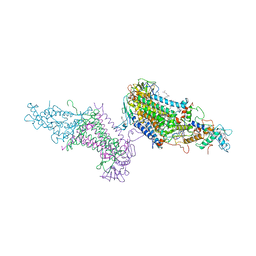 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (QB-DEPLETED) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB.
Structure, 5, 1997
|
|
2Z26
 
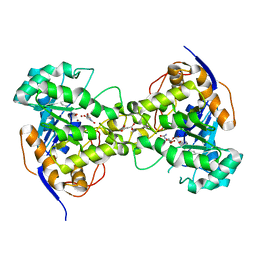 | | Thr110Ala dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, MALONATE ION, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2Z28
 
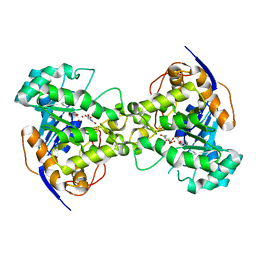 | | Thr109Val dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2Z24
 
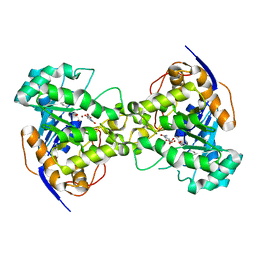 | | Thr110Ser dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2Z2B
 
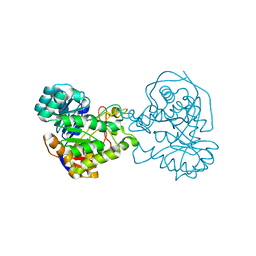 | |
2Z25
 
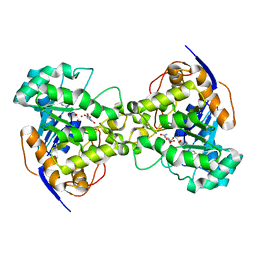 | | Thr110Val dihydroorotase from E. coli | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ... | | Authors: | Lee, M, Maher, M.J, Guss, J.M. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Kinetic and Structural Analysis of Mutant Escherichia coli Dihydroorotases: A Flexible Loop Stabilizes the Transition State
Biochemistry, 46, 2007
|
|
2GZR
 
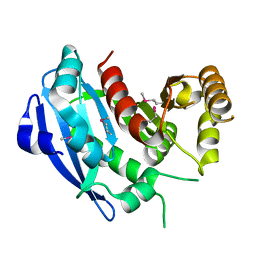 | |
2GJ5
 
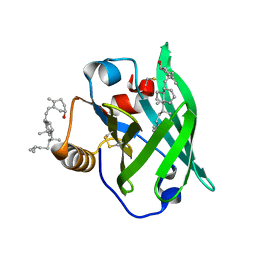 | | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin | | Descriptor: | (1S,3Z)-3-[(2E)-2-[(1R,3AR,7AS)-7A-METHYL-1-[(2R)-6-METHYLHEPTAN-2-YL]-2,3,3A,5,6,7-HEXAHYDRO-1H-INDEN-4-YLIDENE]ETHYLI DENE]-4-METHYLIDENE-CYCLOHEXAN-1-OL, Beta-lactoglobulin | | Authors: | Yang, M.C, Guan, H.H, Liu, M.Y, Yang, J.M, Chen, W.L, Chen, C.J, Mao, S.J. | | Deposit date: | 2006-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a secondary vitamin D3 binding site of milk beta-lactoglobulin.
Proteins, 71, 2008
|
|
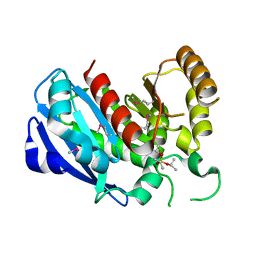
2GZS
 
 | |
2D2E
 
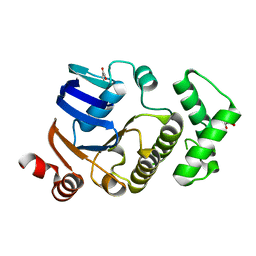 | | Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8 | | Descriptor: | CHLORIDE ION, GLYCEROL, SufC protein | | Authors: | Watanabe, S, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-09-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Atypical Cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8.
J.Mol.Biol., 353, 2005
|
|
2EIO
 
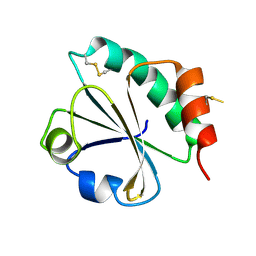 | |
2EIQ
 
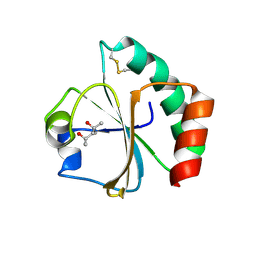 | |
2D2F
 
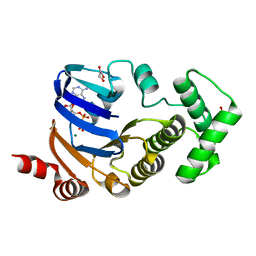 | | Crystal structure of atypical cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Watanabe, S, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-09-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Atypical Cytoplasmic ABC-ATPase SufC from Thermus thermophilus HB8.
J.Mol.Biol., 353, 2005
|
|
3T9G
 
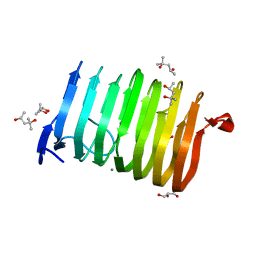 | | The crystal structure of family 3 pectate lyase from Caldicellulosiruptor bescii | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2011-08-02 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A 1.5 A resolution X-ray structure of the catalytic module of Caldicellulosiruptor bescii family 3 pectate lyase.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2FDO
 
 | | Crystal Structure of the Conserved Protein of Unknown Function AF2331 from Archaeoglobus fulgidus DSM 4304 Reveals a New Type of Alpha/Beta Fold | | Descriptor: | Hypothetical protein AF2331 | | Authors: | Wang, S, Kirillova, O, Chruszcz, M, Cymborowski, M.T, Skarina, T, Gorodichtchenskaia, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AF2331 protein from Archaeoglobus fulgidus DSM 4304 forms an unusual interdigitated dimer with a new type of alpha + beta fold.
Protein Sci., 18, 2009
|
|
