103M
 
 | | SPERM WHALE MYOGLOBIN H64A N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-16 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
106M
 
 | | SPERM WHALE MYOGLOBIN V68F ETHYL ISOCYANIDE AT PH 9.0 | | Descriptor: | ETHYL ISOCYANIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-21 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
107M
 
 | | SPERM WHALE MYOGLOBIN V68F N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
108M
 
 | | SPERM WHALE MYOGLOBIN V68F N-BUTYL ISOCYANIDE AT PH 7.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-23 | | Release date: | 1998-05-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
109M
 
 | | SPERM WHALE MYOGLOBIN D122N ETHYL ISOCYANIDE AT PH 9.0 | | Descriptor: | ETHYL ISOCYANIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-22 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
110M
 
 | | SPERM WHALE MYOGLOBIN D122N METHYL ISOCYANIDE AT PH 9.0 | | Descriptor: | METHYL ISOCYANIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-23 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
4WCH
 
 | | Structure of Isolated D Chain of Gigant Hemoglobin from Glossoscolex paulistus | | Descriptor: | Isolated Chain D of Gigant Hemoglobin from Glossoscolex Paulistus, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bachega, J.F.R, Maluf, F.V, Pereira, H.M, Brandao-Neto, J, Tabak, M, Garratt, R.C, Horjales, E. | | Deposit date: | 2014-09-04 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of the giant haemoglobin from Glossoscolex paulistus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WCM
 
 | |
112M
 
 | | SPERM WHALE MYOGLOBIN D122N N-PROPYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-PROPYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-24 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
6LJR
 
 | | human galectin-16 R55N/H57R | | Descriptor: | Galectin-16, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human galectin-16 has a pseudo ligand binding site and plays a role in regulating c-Rel-mediated lymphocyte activity.
Biochim Biophys Acta Gen Subj, 1865, 2020
|
|
2PQC
 
 | | CP4 EPSPS liganded with (R)-phosphonate tetrahedral reaction intermediate analog | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, [3R-[3A,4A,5B(R*)]]-5-(1-CARBOXY-1-PHOSPHONOETHOXY)-4-HYDROXY-3-(PHOSPHONOOXY)-1-CYCLOHEXENE-1-CARBOXYLIC ACID | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
2PQB
 
 | | CP4 EPSPS liganded with (R)-difluoromethyl tetrahedral intermediate analog | | Descriptor: | (3R,4S,5R)-5-[(1R)-1-CARBOXY-2,2-DIFLUORO-1-(PHOSPHONOOXY)ETHOXY]-4-HYDROXY-3-(PHOSPHONOOXY)CYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Healy-Fried, M.L, Funke, T, Han, H, Schonbrunn, E. | | Deposit date: | 2007-05-01 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Differential inhibition of class I and class II 5-enolpyruvylshikimate-3-phosphate synthases by tetrahedral reaction intermediate analogues.
Biochemistry, 46, 2007
|
|
5LLF
 
 | |
4TVT
 
 | | New ligand for thaumatin discovered using acoustic high throughput screening | | Descriptor: | ASCORBIC ACID, L(+)-TARTARIC ACID, SODIUM ION, ... | | Authors: | Teplitsky, E, Joshi, K, Ericson, D.L, Scalia, A, Mullen, J.D, Sweet, R.M, Soares, A.S. | | Deposit date: | 2014-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High throughput screening using acoustic droplet ejection to combine protein crystals and chemical libraries on crystallization plates at high density.
J.Struct.Biol., 191, 2015
|
|
4XAQ
 
 | | mGluR2 ECD and mGluR3 ECD with ligands | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, CHLORIDE ION, Metabotropic glutamate receptor 2, ... | | Authors: | Clawson, D.K. | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-Disubstituted Analogs of 1S,2S,5R,6S-2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: Identification of a Potent, Selective Metabotropic Glutamate Receptor Agonist and Determination of Agonist-Bound Human mGlu2 and mGlu3 Amino Terminal Domain Structures.
J.Med.Chem., 58, 2015
|
|
4WCL
 
 | |
1ANR
 
 | |
5LDB
 
 | | Crystal Structure of Polyphosphate Kinase from Meiothermus ruber bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gerhardt, S, Einsle, O, Kemper, F, Schwarzer, N. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate recognition and mechanism revealed by ligand-bound polyphosphate kinase 2 structures.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4WCN
 
 | | Crystal Structure of Tripeptide bound Cell Shape Determinant Csd4 protein from Helicobacter pylori | | Descriptor: | Conserved hypothetical secreted protein, IODIDE ION, N-acetyl-L-alanyl-N-[(1S,5R)-5-amino-1,5-dicarboxypentyl]-D-glutamine, ... | | Authors: | Chan, A.C, Murphy, M.E. | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Helical Shape of Helicobacter pylori Requires an Atypical Glutamine as a Zinc Ligand in the Carboxypeptidase Csd4.
J.Biol.Chem., 290, 2015
|
|
4WCK
 
 | |
6U4X
 
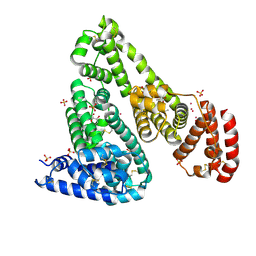 | | Crystal structure of Equine Serum Albumin complex with ibuprofen | | Descriptor: | IBUPROFEN, SULFATE ION, Serum albumin, ... | | Authors: | Czub, M.P, Handing, K.B, Venkataramany, B.S, Steen, E.H, Shabalin, I.G, Satchell, K.J, Joachimiak, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-26 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Albumin-Based Transport of Nonsteroidal Anti-Inflammatory Drugs in Mammalian Blood Plasma.
J.Med.Chem., 63, 2020
|
|
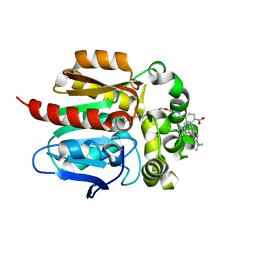
6U32
 
 | |
4XAR
 
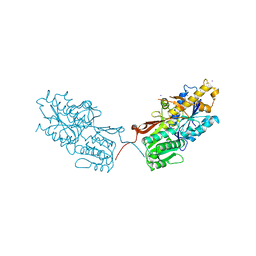 | | mGluR2 ECD and mGluR3 ECD complex with ligands | | Descriptor: | (1S,2S,5R,6S)-2-aminobicyclo[3.1.0]hexane-2,6-dicarboxylic acid, IODIDE ION, Metabotropic glutamate receptor 3 | | Authors: | Clawson, D.K. | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-11 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4-Disubstituted Analogs of 1S,2S,5R,6S-2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylate: Identification of a Potent, Selective Metabotropic Glutamate Receptor Agonist and Determination of Agonist-Bound Human mGlu2 and mGlu3 Amino Terminal Domain Structures.
J.Med.Chem., 58, 2015
|
|
7PKD
 
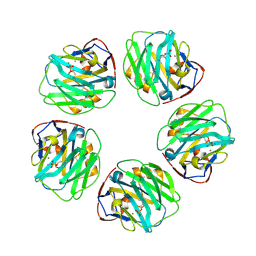 | | C-reactive protein decamer at pH 7.5 with phosphocholine ligand | | Descriptor: | C-reactive protein, CALCIUM ION, PHOSPHOCHOLINE | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
7PKH
 
 | | C-reactive protein decamer at pH 5 with phosphocholine ligand | | Descriptor: | C-reactive protein, CALCIUM ION, PHOSPHOCHOLINE | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2022-01-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
