7P7J
 
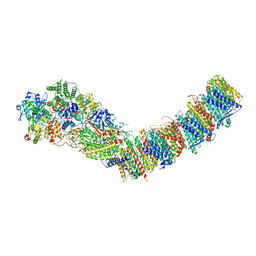 | | Complex I from E. coli, DDM/LMNG-purified, with DQ, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-decyl-5,6-dimethoxy-3-methylcyclohexa-2,5-diene-1,4-dione, CALCIUM ION, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7M
 
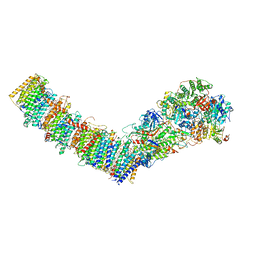 | | Complex I from E. coli, DDM/LMNG-purified, inhibited by Piericidin A, Open state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
7P7K
 
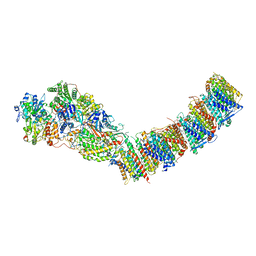 | | Complex I from E. coli, DDM/LMNG-purified, with DQ, Resting state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, EICOSANE, ... | | Authors: | Kravchuk, V, Kampjut, D, Sazanov, L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A universal coupling mechanism of respiratory complex I.
Nature, 609, 2022
|
|
3TOB
 
 | |
7P19
 
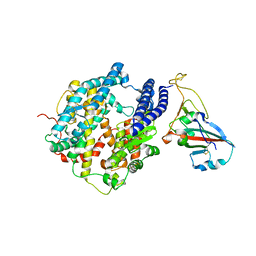 | |
7P0Q
 
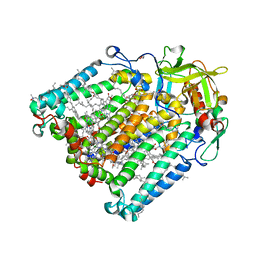 | | F(M197)H mutant structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV by fixed-target serial synchrotron crystallography (100K, 26keV) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Guenther, S, Meents, A, Fufina, T.Y, Vasilieva, L.G. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-Ray structure of Rhodobacter sphaeroides reaction center with M197 Phe-His substitution clarifies properties of the mutant complex
To Be Published
|
|
3U8X
 
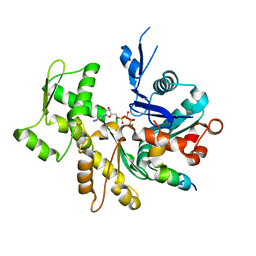 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-29) of drosophila Ciboulot and the C-terminal domain (residues 18-44) of bovine Thymosin-beta4, bound to G-actin-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
3OZO
 
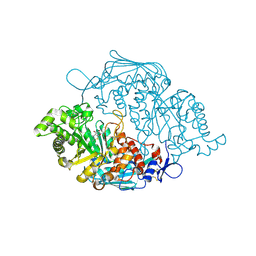 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | Deposit date: | 2010-09-27 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT
To be Published
|
|
7P0W
 
 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing thymine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*TP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
7OXE
 
 | |
3P4Q
 
 | | Crystal structure of Menaquinol:oxidoreductase in complex with oxaloacetate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-06 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
3ONZ
 
 | | Human tetrameric hemoglobin: proximal nitrite ligand at beta | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, NITRITE ION, ... | | Authors: | Yi, J, Thormas, L.M, Safo, M.K, Musayev, F.N, Richter-Addo, G.B. | | Deposit date: | 2010-08-30 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Crystallographic Trapping of Heme Loss Intermediates during the Nitrite-Induced Degradation of Human Hemoglobin.
Biochemistry, 50, 2011
|
|
7OY7
 
 | | Crystal structure of a trapped Pab-AGOG/double-standed DNA covalent intermediate (DNA containing cytosine opposite to lesion) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(PED)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Goffinont, S, Flament, D, Castaing, B. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
3OO5
 
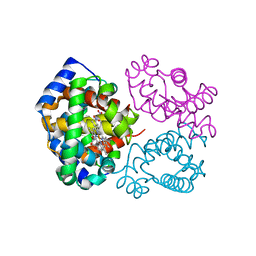 | | R-state human hemoglobin: nitriheme modified | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, NITRITE ION, ... | | Authors: | Yi, J, Thormas, L.M, Richter-Addo, G.B. | | Deposit date: | 2010-08-30 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Trapping of Heme Loss Intermediates during the Nitrite-Induced Degradation of Human Hemoglobin.
Biochemistry, 50, 2011
|
|
7OYE
 
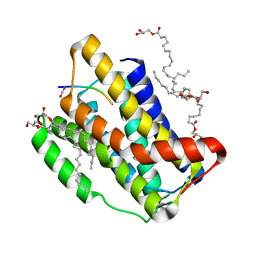 | |
7P17
 
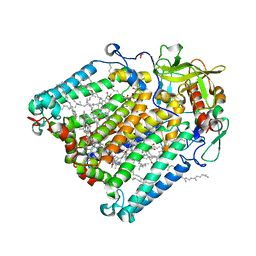 | | F(M197)H mutant structure of Photosynthetic Reaction Center From Rhodobacter Sphaeroides strain RV by fixed-target serial synchrotron crystallography (room temperature, 12keV) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, ... | | Authors: | Gabdulkhakov, A.G, Selikhanov, G.K, Guenther, S, Meents, A, Fufina, T.Y, Vasilieva, L.G. | | Deposit date: | 2021-07-01 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | X-Ray structure of Rhodobacter sphaeroides reaction center with M197 Phe-His substitution clarifies properties of the mutant complex
To Be Published
|
|
3U3U
 
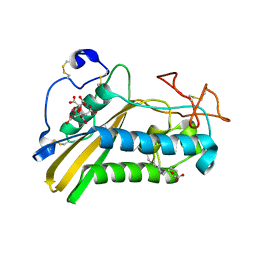 | | Crystal structure of the tablysin-15-leukotriene E4 complex | | Descriptor: | (5S,7E,9E,11Z,14Z)-5-hydroxyicosa-7,9,11,14-tetraenoic acid, CITRIC ACID, PRASEODYMIUM ION, ... | | Authors: | Andersen, J.F. | | Deposit date: | 2011-10-06 | | Release date: | 2012-02-15 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of protein having inhibitory disintegrin and leukotriene scavenging functions contained in single domain.
J.Biol.Chem., 287, 2012
|
|
7P33
 
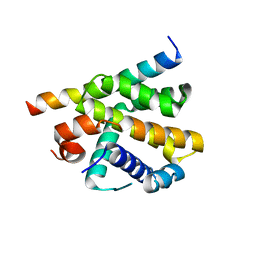 | | Epstein-Barr virus encoded Bcl-2 homolog BHRF-1 in complex with Bid BH3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, BH3-interacting domain death agonist p15, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.78542733 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
3P0B
 
 | |
3P2T
 
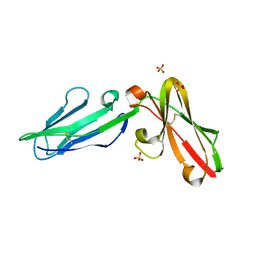 | | Crystal Structure of Leukocyte Ig-like Receptor LILRB4 (ILT3/LIR-5/CD85k) | | Descriptor: | Leukocyte immunoglobulin-like receptor subfamily B member 4, SULFATE ION | | Authors: | Chen, Y, Nam, G, Cheng, H, Zhang, J.H, Willcox, B.E, Gao, G.F. | | Deposit date: | 2010-10-04 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure of leukocyte Ig-like receptor LILRB4 (ILT3/LIR-5/CD85k): a myeloid inhibitory receptor involved in immune tolerance
J.Biol.Chem., 286, 2011
|
|
3TYO
 
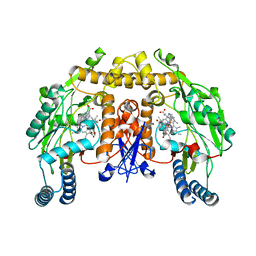 | | Structure of neuronal nitric oxide synthase heme domain in complex with 6-(((3S,4S)-4-(2-((furan-2-ylmethyl)amino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3S,4S)-4-{2-[(furan-2-ylmethyl)amino]ethoxy}pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2011-09-26 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.927 Å) | | Cite: | Intramolecular hydrogen bonding: A potential strategy for more bioavailable inhibitors of neuronal nitric oxide synthase.
Bioorg.Med.Chem., 20, 2012
|
|
3TLT
 
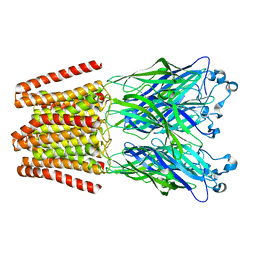 | | The GLIC pentameric Ligand-Gated Ion Channel H11'F mutant in a locally-closed conformation (LC1 subtype) | | Descriptor: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Glr4197 protein | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-08-30 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3P4R
 
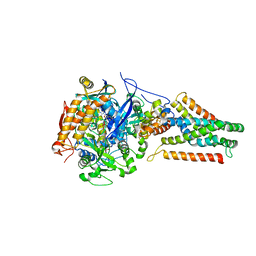 | | Crystal structure of Menaquinol:fumarate oxidoreductase in complex with glutarate | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Archuleta, T.L, Andrell, J, Luna-Chavez, C, Davis, T.A, Sarwar, M, Ham, A.J, McDonald, W.H, Yankowskaya, V, Stern, H.A, Johnston, J.N, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2010-10-07 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Geometric restraint drives on- and off-pathway catalysis by the Escherichia coli menaquinol:fumarate reductase.
J.Biol.Chem., 286, 2011
|
|
7P8L
 
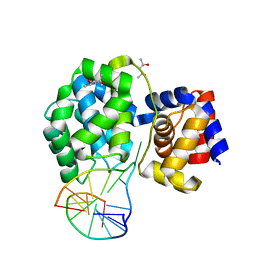 | | Crystal structure of Pyrococcus abyssi 8-oxoguanine DNA glycosylase (PabAGOG) in complex with dsDNA containing cytosine opposite to 8-oxoG | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*AP*GP*AP*AP*AP*CP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*(8OG)P*TP*TP*TP*CP*T)-3'), ... | | Authors: | Coste, F, Flament, D, Castaing, B. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
3P97
 
 | |
