7P7Q
 
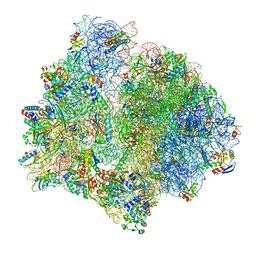 | | E. faecalis 70S ribosome bound by PoxtA-EQ2, high-resolution combined volume | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7P7S
 
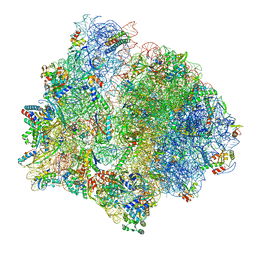 | | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state II | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7P7U
 
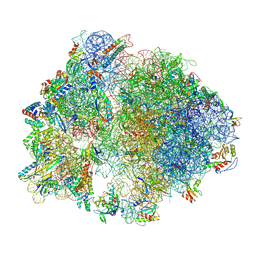 | | E. faecalis 70S ribosome with P-tRNA, state IV | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
7P7R
 
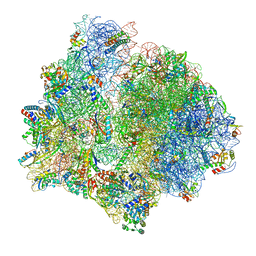 | | PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state I | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Wilson, D.N. | | Deposit date: | 2021-07-20 | | Release date: | 2022-03-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for PoxtA-mediated resistance to phenicol and oxazolidinone antibiotics.
Nat Commun, 13, 2022
|
|
1OW5
 
 | |
1OYX
 
 | | CRYSTAL STRUCTURE OF 3-MBT REPEATS OF LETHAL (3) MALIGNANT BRAIN TUMOR (SELENO-MET) AT 1.85 ANGSTROM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Wang, W.K, Tereshko, V, Boccuni, P, MacGrogan, D, Nimer, S.D, Patel, D.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Structure, 11, 2003
|
|
1OZ2
 
 | | CRYSTAL STRUCTURE OF 3-MBT REPEATS OF LETHAL (3) MALIGNANT BRAIN TUMOR (NATIVE-II) AT 1.55 ANGSTROM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Wang, W.K, Tereshko, V, Boccuni, P, MacGrogan, D, Nimer, S.D, Patel, D.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Structure, 11, 2003
|
|
1OZ3
 
 | | Crystal Structure of 3-MBT repeats of lethal (3) malignant Brain Tumor (Native-I) at 1.85 angstrom | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Wang, W.K, Tereshko, V, Boccuni, P, MacGrogan, D, Nimer, S.D, Patel, D.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Structure, 11, 2003
|
|
3ZF8
 
 | |
5GO2
 
 | | Crystal structure of chorismate mutase like domain of bifunctional DAHP synthase of Bacillus subtilis in complex with Citrate | | Descriptor: | CITRIC ACID, Protein AroA(G), SULFATE ION | | Authors: | Pratap, S, Dev, A, Sharma, V, Yadav, R, Narwal, M, Tomar, S, Kumar, P. | | Deposit date: | 2016-07-26 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | Structure of Chorismate Mutase-like Domain of DAHPS from Bacillus subtilis Complexed with Novel Inhibitor Reveals Conformational Plasticity of Active Site.
Sci Rep, 7, 2017
|
|
5GMU
 
 | | Crystal structure of chorismate mutase like domain of bifunctional DAHP synthase of Bacillus subtilis in complex with Chlorogenic acid | | Descriptor: | (1R,3R,4S,5R)-3-[3-[3,4-bis(oxidanyl)phenyl]propanoyloxy]-1,4,5-tris(oxidanyl)cyclohexane-1-carboxylic acid, Protein AroA(G), SULFATE ION | | Authors: | Pratap, S, Dev, A, Sharma, V, Yadav, R, Narwal, M, Tomar, S, Kumar, P. | | Deposit date: | 2016-07-16 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Chorismate Mutase-like Domain of DAHPS from Bacillus subtilis Complexed with Novel Inhibitor Reveals Conformational Plasticity of Active Site.
Sci Rep, 7, 2017
|
|
4WSC
 
 | | Crystal structure of a GroELK105A mutant | | Descriptor: | 60 kDa chaperonin | | Authors: | Lorimer, G.H, Ye, X, Fei, X, Yang, D, Corsepius, N, LaRonde, N.A. | | Deposit date: | 2014-10-26 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of a GroELK105A mutant
To Be Published
|
|
7CD7
 
 | | GFP-40/GFPuv complex, Form I | | Descriptor: | GFP-40, Green fluorescent protein | | Authors: | Yasui, N, Yamashita, A. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | A sweet protein monellin as a non-antibody scaffold for synthetic binding proteins.
J.Biochem., 169, 2021
|
|
7CD8
 
 | | GFP-40/GFPuv complex, Form II | | Descriptor: | GFP-40, Green fluorescent protein | | Authors: | Yasui, N, Yamashita, A. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A sweet protein monellin as a non-antibody scaffold for synthetic binding proteins.
J.Biochem., 169, 2021
|
|
3QVN
 
 | |
4APQ
 
 | | Crystal structure of autoreactive-Valpha14-Vbeta6 NKT TCR in complex with CD1d-sulfatide | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIGEN-PRESENTING GLYCOPROTEIN CD1D1, ... | | Authors: | Clarke, A.J, Le Nours, J, Rossjohn, J. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Type-II Natural Killer T Cell Antigen Receptor Mediated Recognition of Cd1D-Sulfatide
To be Published
|
|
3KXY
 
 | | Crystal Structure of the ExsC-ExsE Complex | | Descriptor: | Exoenzyme S synthesis protein C, ExsE | | Authors: | Vogelaar, N.J, Robinson, H.H, Schubot, F.D. | | Deposit date: | 2009-12-04 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Analysis of the Crystal Structure of the ExsC.ExsE Complex Reveals Distinctive Binding Interactions of the Pseudomonas aeruginosa Type III Secretion Chaperone ExsC with ExsE and ExsD.
Biochemistry, 49, 2010
|
|
6S1L
 
 | |
6S1R
 
 | |
6S29
 
 | |
8J7C
 
 | | Crystal structure of triosephosphate isomerase from Leishmania orientalis at 1.88A with an arsenic ion bound at Cys57 | | Descriptor: | ARSENIC, Triosephosphate isomerase | | Authors: | Kuaprasert, B, Attarataya, J, Riangrungroj, P, Pornthanakasem, W, Suginta, W, Mungthin, M, Leelayoova, S, Choowongkomon, K, Leartsakulpanich, U. | | Deposit date: | 2023-04-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Leishmania orientalis triosephosphate isomerase crystal structure at 1.45 angstroms resolution and its potential specific inhibitors
To be published
|
|
8JBP
 
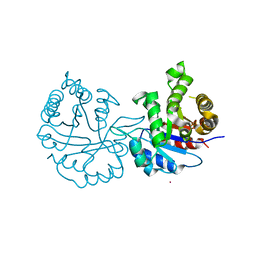 | | Crystal structure of triosephosphate isomerase from Leishmania orientalis at 1.45 angstroms resolution with an arsenic atom bound at Cys57 | | Descriptor: | ARSENIC, Triosephosphate isomerase | | Authors: | Kuaprasert, B, Leartsakulpanich, U, Riangrungroj, P, Pornthanakasem, W, Suginta, W, Robinson, R.C, Zhou, Y, Mungthin, M, Leelayoova, S, Saehlee, S, Choowongkomon, K. | | Deposit date: | 2023-05-09 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Leishmania orientalis triosephosphate isomerase crystal structure at 1.45 angstroms resolution and its potential specific inhibitors
To be published
|
|
3S6I
 
 | |
8OEQ
 
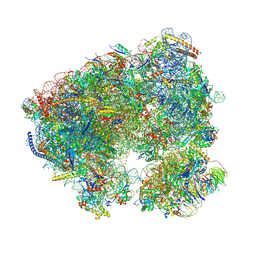 | | Crystal structure of the Candida albicans 80S ribosome in complex with Paromomycin (250uM) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-12 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
2HK3
 
 | |
