3ZBY
 
 | | Ligand-free structure of CYP142 from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Garcia-Fernandez, E, Frank, D.J, Galan, B, Kells, P.M, Podust, L.M, Garcia, J.L, Ortiz de Montellano, P.R. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
7YJF
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268P/V78I in complex with N-imidazolyl-pentanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, HYDROXYAMINE, P450 BM3 heme domain mutant F87A/T268P/V78I, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-20 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7YDD
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268P/V78I in complex with N-imidazolyl-pentanoyl-L-phenylalanine,propylbenzene and hydroxylamine | | Descriptor: | (2~{S})-2-(5-imidazol-1-ylpentanoylamino)-3-phenyl-propanoic acid, HYDROXYAMINE, P450 BM3 heme domain mutant F87A/T268P/V78I, ... | | Authors: | Dong, S, Chen, J, Jiang, Y, Cong, Z, Feng, Y. | | Deposit date: | 2022-07-04 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.663 Å) | | Cite: | Regiodivergent and Enantioselective Hydroxylation of C-H bonds by Synergistic Use of Protein Engineering and Exogenous Dual-Functional Small Molecules.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3BUJ
 
 | | Crystal Structure of CalO2 | | Descriptor: | CalO2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McCoy, J.G, Johnson, H.D, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-02 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural characterization of CalO2: a putative orsellinic acid P450 oxidase in the calicheamicin biosynthetic pathway.
Proteins, 74, 2009
|
|
2YOO
 
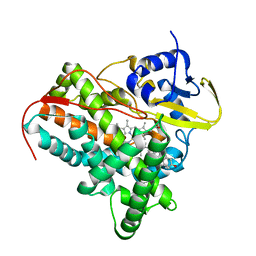 | | Cholest-4-en-3-one bound structure of CYP142 from Mycobacterium smegmatis | | Descriptor: | (8ALPHA,9BETA)-CHOLEST-4-EN-3-ONE, MAGNESIUM ION, P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Garcia-Fernandez, E, Frank, D.J, Galan, B, Kells, P.M, Podust, L.M, Garcia, J.L, Ortiz de Montellano, P.R. | | Deposit date: | 2012-10-25 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
3WRM
 
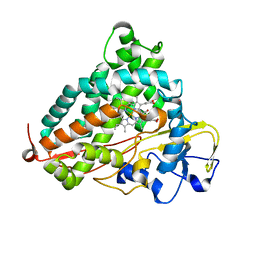 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of P450cam intermedite
To be published
|
|
3WRJ
 
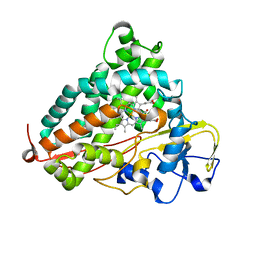 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of P450cam intermedite
to be published
|
|
3WRL
 
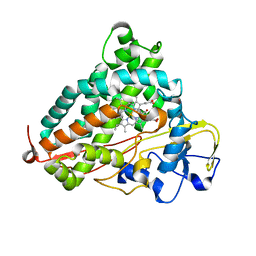 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of P450cam intermedite
To be published
|
|
1E6E
 
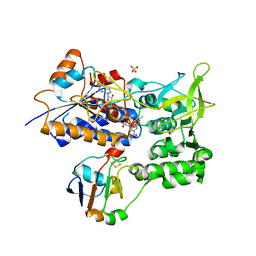 | | ADRENODOXIN REDUCTASE/ADRENODOXIN COMPLEX OF MITOCHONDRIAL P450 SYSTEMS | | Descriptor: | ADRENODOXIN, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mueller, J.J, Lapko, A, Bourenkov, G, Ruckpaul, K, Heinemann, U. | | Deposit date: | 2000-08-15 | | Release date: | 2001-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adrenodoxin Reductase-Adrenodoxin Complex Structure Suggests Electron Transfer Path in Steroid Biosynthesis.
J.Biol.Chem., 276, 2001
|
|
4C9O
 
 | |
3HUI
 
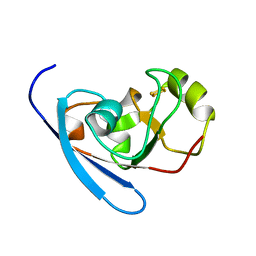 | | Crystal Structure of the mutant A105R of [2Fe-2S] Ferredoxin in the Class I CYP199A2 System from Rhodopseudomonas palustris | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Bell, S.G, Xu, F, Rao, Z, Wong, L.-L. | | Deposit date: | 2009-06-14 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Protein recognition in ferredoxin-P450 electron transfer in the class I CYP199A2 system from Rhodopseudomonas palustris
J.Biol.Inorg.Chem., 15, 2010
|
|
1YJJ
 
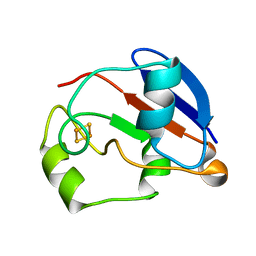 | | RDC-refined Solution NMR structure of oxidized putidaredoxin | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Jain, N.U, Tjioe, E, Savidor, A, Boulie, J. | | Deposit date: | 2005-01-14 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Redox-dependent structural differences in putidaredoxin derived from homologous structure refinement via residual dipolar couplings.
Biochemistry, 44, 2005
|
|
4C9P
 
 | |
4C9L
 
 | |
4C9N
 
 | |
4C9K
 
 | |
8AMP
 
 | | Crystal structure of M.tuberculosis ferredoxin Fdx | | Descriptor: | FE (III) ION, FE3-S4 CLUSTER, Possible ferredoxin | | Authors: | Bukhdruker, S, Kavaleuski, A, Marin, E, Kapranov, I, Mishin, A, Gilep, A, Strushkevich, N, Borshchevskiy, V. | | Deposit date: | 2022-08-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into 3Fe-4S ferredoxins diversity in M. tuberculosis highlighted by a first redox complex with P450.
Front Mol Biosci, 9, 2022
|
|
1CJC
 
 | |
4OXX
 
 | |
4APY
 
 | | Ethylene glycol-bound form of P450 CYP125A3 from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, P450 HEME-THIOLATE PROTEIN, ... | | Authors: | Frank, D.J, Garcia Fernandez, E, Kells, P.M, Garcia Lopez, J.L, Podust, L.M, Ortiz de Montellano, P.R. | | Deposit date: | 2012-04-09 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Highly Conserved Mycobacterial Cholesterol Catabolic Pathway.
Environ.Microbiol., 15, 2013
|
|
1OQQ
 
 | | Crystal structure of C73S/C85S mutant of putidaredoxin, a [2Fe-2S] ferredoxin from Pseudomonas putida, at 1.47A resolution | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Sevrioukova, I.F, Garcia, C, Li, H, Bhaskar, B, Poulos, T.L. | | Deposit date: | 2003-03-10 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal structure of putidaredoxin, the [2Fe-2S] component of the P450cam monooxygenase system from Pseudomonas putida
J.MOL.BIOL., 333, 2003
|
|
1OQR
 
 | | Crystal structure of C73S mutant of putidaredoxin, a [2Fe-2S] ferredoxin from Pseudomonas putida, at 1.65A resolution | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Sevrioukova, I.F, Garcia, C, Li, H, Bhaskar, B, Poulos, T.L. | | Deposit date: | 2003-03-10 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of putidaredoxin, the [2Fe-2S] component of the P450cam monooxygenase system from Pseudomonas putida
J.MOL.BIOL., 333, 2003
|
|
2YP1
 
 | | Crystallization of a 45 kDa peroxygenase- peroxidase from the mushroom Agrocybe aegerita and structure determination by SAD utilizing only the haem iron | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Piontek, K, Strittmatter, E, Ullrich, R, Plattner, D.A, Hofrichter, M. | | Deposit date: | 2012-10-29 | | Release date: | 2013-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Basis of Substrate Conversion in a New Aromatic Peroxygenase: P450 Functionality with Benefits
J.Biol.Chem., 288, 2013
|
|
2YOR
 
 | | Crystallization of a 45 kDa peroxygenase- peroxidase from the mushroom Agrocybe aegerita and structure determination by SAD utilizing only the haem iron | | Descriptor: | 1H-imidazol-5-ylmethanol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Piontek, K, Strittmatter, E, Ullrich, R, Plattner, D.A, Hofrichter, M. | | Deposit date: | 2012-10-26 | | Release date: | 2013-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis of Substrate Conversion in a New Aromatic Peroxygenase: P450 Functionality with Benefits
J.Biol.Chem., 288, 2013
|
|
9BBB
 
 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, PROTOPORPHYRIN IX CONTAINING FE, cobicistat | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2024-04-05 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interaction of CYP3A4 with the inhibitor cobicistat: Structural and mechanistic insights and comparison with ritonavir
to be published
|
|
