8HAT
 
 | |
5XUY
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
3KCS
 
 | | Crystal structure of PAmCherry1 in the dark state | | Descriptor: | PAmCherry1 protein | | Authors: | Malashkevich, V.N, Subach, F.V, Zencheck, W.D, Xiao, H, Filonov, G.S, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KCT
 
 | | CRYSTAL STRUCTURE OF PAmCherry1 in the photoactivated state | | Descriptor: | PAmCherry1 protein | | Authors: | Malashkevich, V.N, Subach, F.V, Zencheck, W.D, Xiao, H, Filonov, G.S, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3FZA
 
 | | Crystal structure of poplar glutaredoxin S12 in complex with glutathione and beta-mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, GLUTATHIONE, Glutaredoxin | | Authors: | Didierjean, C, Corbier, C, Koh, C.S, Rouhier, N, Jacquot, J.P. | | Deposit date: | 2009-01-24 | | Release date: | 2009-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site.
J.Biol.Chem., 284, 2009
|
|
3FZ9
 
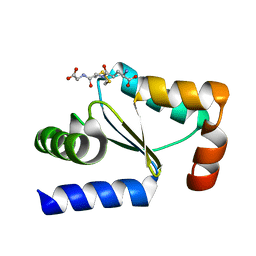 | | Crystal structure of poplar glutaredoxin S12 in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutaredoxin | | Authors: | Didierjean, C, Corbier, C, Koh, C.S, Rouhier, N, Jacquot, J.P. | | Deposit date: | 2009-01-24 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site.
J.Biol.Chem., 284, 2009
|
|
3JBM
 
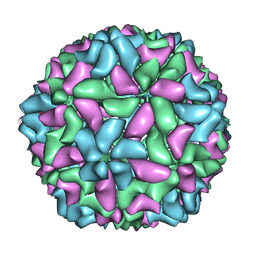 | | Electron cryo-microscopy of a virus-like particle of orange-spotted grouper nervous necrosis virus | | Descriptor: | virus-like particle of orange-spotted grouper nervous necrosis virus | | Authors: | Xie, J, Li, K, Gao, Y, Huang, R, Lai, Y, Shi, Y, Yang, S, Zhu, G, Zhang, Q, He, J. | | Deposit date: | 2015-09-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural analysis and insertion study reveal the ideal sites for surface displaying foreign peptides on a betanodavirus-like particle
Vet. Res., 47, 2016
|
|
3IR8
 
 | | Red fluorescent protein mKeima at pH 7.0 | | Descriptor: | Large stokes shift fluorescent protein | | Authors: | Henderson, J.N, Osborn, M.F, Koon, N, Gepshtein, R, Huppert, D, Remington, S.J. | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Excited state proton transfer in the red fluorescent protein mKeima.
J.Am.Chem.Soc., 131, 2009
|
|
5K4Z
 
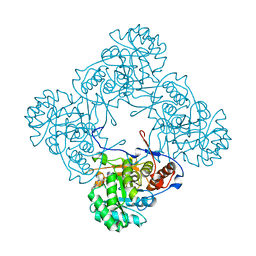 | | M. thermoresistible IMPDH in complex with IMP and Compound 6 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(4-fluorophenyl)-4-(2~{H}-indazol-6-ylsulfamoyl)-3,5-dimethyl-1~{H}-pyrrole-2-carboxamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
5K4X
 
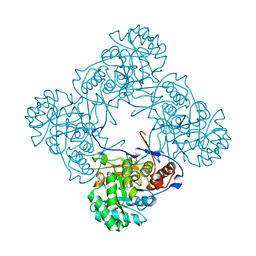 | | M. thermoresistible IMPDH in complex with IMP and Compound 1 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(2~{H}-indazol-6-yl)-3,5-dimethyl-1~{H}-pyrazole-4-sulfonamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
5LTQ
 
 | |
5LTR
 
 | |
5LU3
 
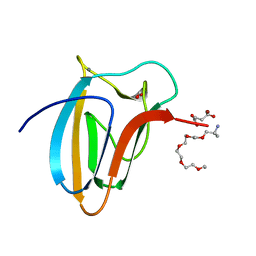 | | The Structure of Spirochaeta thermophila CBM64 | | Descriptor: | 3,6,9,12,15-pentaoxaoctadecan-17-amine, 4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Correia, M.A.S, Romao, M.J, Carvalho, A.L. | | Deposit date: | 2016-09-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stability and Ligand Promiscuity of Type A Carbohydrate-binding Modules Are Illustrated by the Structure of Spirochaeta thermophila StCBM64C.
J. Biol. Chem., 292, 2017
|
|
5LTP
 
 | |
2VMA
 
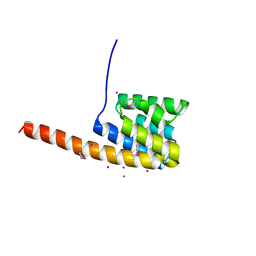 | | The three-dimensional structure of the cytoplasmic domains of EpsF from the Type 2 Secretion System of Vibrio cholerae | | Descriptor: | CALCIUM ION, GENERAL SECRETION PATHWAY PROTEIN F, IODIDE ION | | Authors: | Abendroth, J, Korotkov, K.V, Mitchell, D.D, Kreger, A, Hol, W.G.J. | | Deposit date: | 2008-01-25 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Three-Dimensional Structure of the Cytoplasmic Domains of Epsf from the Type 2 Secretion System of Vibrio Cholerae.
J.Struct.Biol., 166, 2009
|
|
2XVY
 
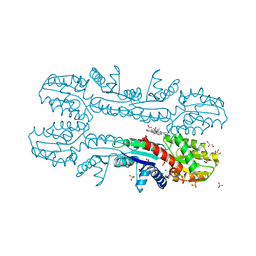 | | Cobalt chelatase CbiK (periplasmic) from Desulvobrio vulgaris Hildenborough (co-crystallised with cobalt and SHC) | | Descriptor: | CHELATASE, PUTATIVE, COBALT (II) ION, ... | | Authors: | Romao, C.V, Lobo, S.A.L, Carrondo, M.A, Saraiva, L.M, Matias, P.M. | | Deposit date: | 2010-10-28 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Desulfovibrio vulgaris CbiK(P) cobaltochelatase: evolution of a haem binding protein orchestrated by the incorporation of two histidine residues.
Environ. Microbiol., 19, 2017
|
|
2W9Y
 
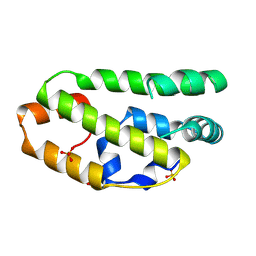 | | The structure of the lipid binding protein Ce-FAR-7 from Caenorhabditis elegans | | Descriptor: | FATTY ACID/RETINOL BINDING PROTEIN PROTEIN 7, ISOFORM A, CONFIRMED BY TRANSCRIPT EVIDENCE, ... | | Authors: | Jordanova, R, Groves, M.R, Tucker, P.A. | | Deposit date: | 2009-01-30 | | Release date: | 2009-10-20 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fatty Acid and Retinoid Binding Proteins Have Distinct Binding Pockets for the Two Types of Cargo
J.Biol.Chem., 284, 2009
|
|
1G7K
 
 | | CRYSTAL STRUCTURE OF DSRED, A RED FLUORESCENT PROTEIN FROM DISCOSOMA SP. RED | | Descriptor: | FLUORESCENT PROTEIN FP583 | | Authors: | Yarbrough, D, Wachter, R.M, Kallio, K, Matz, M.V, Remington, S.J. | | Deposit date: | 2000-11-10 | | Release date: | 2000-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8PMI
 
 | | Structure of Nal1 indica cultivar IR64, construct 36-458 in presence of peptide from FZP protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AMMONIUM ION, MAGNESIUM ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PMG
 
 | | Structure of Nal1 indica cultivar IR64, construct 36-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PML
 
 | | Structure of Nal1 protein , SPIKE allele from japonica rice, construct 46-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
3QAY
 
 | | Catalytic domain of CD27L endolysin targeting Clostridia Difficile | | Descriptor: | Endolysin, PHOSPHATE ION, ZINC ION | | Authors: | Mayer, M.J, Garefaliki, V, Spoerl, R, Narbad, A, Meijers, R. | | Deposit date: | 2011-01-12 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range.
J.Bacteriol., 193, 2011
|
|
5Z6Y
 
 | | Structure of sfYFP48S95C66BPA | | Descriptor: | Green fluorescent protein | | Authors: | Wang, J.Y, Wang, J.Y. | | Deposit date: | 2018-01-25 | | Release date: | 2019-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | structure of sfYFP48S95C66BPA at 1.95 Angstroms resolution
To Be Published
|
|
8PN2
 
 | | CryoEM structure of Nal1 protein, allele IR64, from Oryza sativa indica cultivar | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PMM
 
 | | Structure of Nal1 protein, allele SPIKE from japonica rice, construct 31-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
