1MUV
 
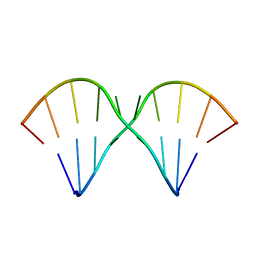 | | Sheared A(anti)-A(anti) Base Pairs in a Destabilizing 2x2 Internal Loop: The NMR Structure of 5'(rGGCAAGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*AP*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Schroeder, S.J, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sheared Aanti-Aanti Base Pairs in a Destabilizing 2x2 Internal Loop: The NMR Structure of
5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
6A7Y
 
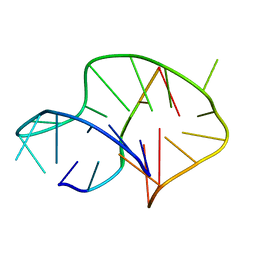 | | Solution structure of an intermolecular leaped V-shape G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*TP*TP*TP*TP*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3'), DNA (5'-D(*TP*GP*GP*GP*GP*A)-3') | | Authors: | Wan, C.J, Zhang, N. | | Deposit date: | 2018-07-05 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an asymmetric intermolecular leaped V-shape G-quadruplex: selective recognition of the d(G2NG3NG4) sequence motif by a short linear G-rich DNA probe.
Nucleic Acids Res., 47, 2019
|
|
1OEF
 
 | | PEPTIDE OF HUMAN APOE RESIDUES 263-286, NMR, 5 STRUCTURES AT PH 4.8, 37 DEGREES CELSIUS AND PEPTIDE:SDS MOLE RATIO OF 1:90 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Wang, G, Pierens, G.K, Treleaven, W.D, Sparrow, J.T, Cushley, R.J. | | Deposit date: | 1996-03-16 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformations of human apolipoprotein E(263-286) and E(267-289) in aqueous solutions of sodium dodecyl sulfate by CD and 1H NMR.
Biochemistry, 35, 1996
|
|
1BMX
 
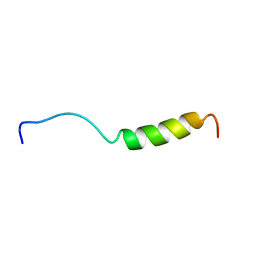 | |
1GPS
 
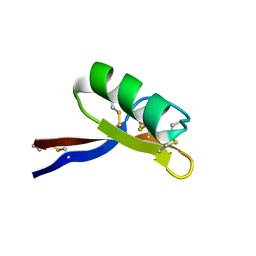 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | Descriptor: | GAMMA-1-P THIONIN | | Authors: | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | Deposit date: | 1992-07-29 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
1OEG
 
 | | PEPTIDE OF HUMAN APOE RESIDUES 267-289, NMR, 5 STRUCTURES AT PH 6.0, 37 DEGREES CELSIUS AND PEPTIDE:SDS MOLE RATIO OF 1:90 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Wang, G, Pierens, G.K, Treleaven, W.D, Sparrow, J.T, Cushley, R.J. | | Deposit date: | 1996-03-16 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformations of human apolipoprotein E(263-286) and E(267-289) in aqueous solutions of sodium dodecyl sulfate by CD and 1H NMR.
Biochemistry, 35, 1996
|
|
1GPT
 
 | | SOLUTION STRUCTURE OF GAMMA 1-H AND GAMMA 1-P THIONINS FROM BARLEY AND WHEAT ENDOSPERM DETERMINED BY 1H-NMR: A STRUCTURAL MOTIF COMMON TO TOXIC ARTHROPOD PROTEINS | | Descriptor: | GAMMA-1-H THIONIN | | Authors: | Bruix, M, Jimenez, M.A, Santoro, J, Gonzalez, C, Colilla, F.J, Mendez, E, Rico, M. | | Deposit date: | 1992-07-29 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of gamma 1-H and gamma 1-P thionins from barley and wheat endosperm determined by 1H-NMR: a structural motif common to toxic arthropod proteins.
Biochemistry, 32, 1993
|
|
1Q7O
 
 | | Determination of f-MLF-OH Peptide Structure with solid-state magic-angle spinning NMR Spectroscopy | | Descriptor: | chemotactic peptide | | Authors: | Rienstra, C.M, Tucker-Kellogg, L, Jaroniec, C.P, Hohwy, M, Reif, B, McMahon, M.T, Tidor, B, Lozano-Perez, T, Griffin, R.G. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-09 | | Last modified: | 2022-03-02 | | Method: | SOLID-STATE NMR | | Cite: | De novo determination of peptide structure with solid-state magic-angle spinning NMR Spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1OLG
 
 | |
1ODQ
 
 | |
1MIC
 
 | |
1ODP
 
 | |
1ODR
 
 | |
3ALC
 
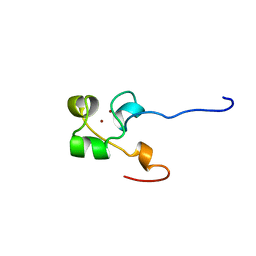 | | ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR DNA-BINDING DOMAIN FROM ASPERGILLUS NIDULANS | | Descriptor: | PROTEIN (ETHANOL REGULON TRANSCRIPTIONAL ACTIVATOR), ZINC ION | | Authors: | Cerdan, R, Cahuzac, B, Felenbok, B, Guittet, E. | | Deposit date: | 1999-03-11 | | Release date: | 2000-05-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of AlcR (1-60) provides insight in the unusual DNA binding properties of this zinc binuclear cluster protein.
J.Mol.Biol., 295, 2000
|
|
1EQ8
 
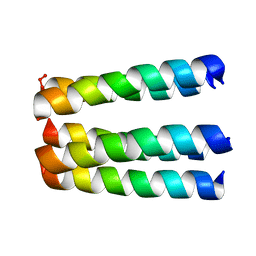 | | THREE-DIMENSIONAL STRUCTURE OF THE PENTAMERIC HELICAL BUNDLE OF THE ACETYLCHOLINE RECEPTOR M2 TRANSMEMBRANE SEGMENT | | Descriptor: | ACETYLCHOLINE RECEPTOR PROTEIN | | Authors: | Marassi, F.M, Gesell, J.J, Kim, Y, Valente, A.P, Oblatt-Montal, M, Montal, M, Opella, S.J. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-26 | | Last modified: | 2024-09-25 | | Method: | SOLID-STATE NMR | | Cite: | Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy.
Nat.Struct.Biol., 6, 1999
|
|
1OLH
 
 | |
1MR0
 
 | | SOLUTION NMR STRUCTURE OF AGRP(87-120; C105A) | | Descriptor: | AGOUTI RELATED PROTEIN | | Authors: | Jackson, P.J, Mcnulty, J.C, Yang, Y.K, Thompson, D.A, Chai, B, Gantz, I, Barsh, G.S, Millhauser, G.M. | | Deposit date: | 2002-09-17 | | Release date: | 2002-10-02 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Design, pharmacology, and NMR structure of a minimized cystine knot with agouti-related protein activity.
Biochemistry, 41, 2002
|
|
1G2T
 
 | |
1EZA
 
 | |
1EZB
 
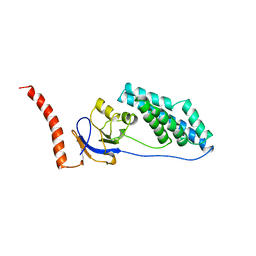 | |
1G2S
 
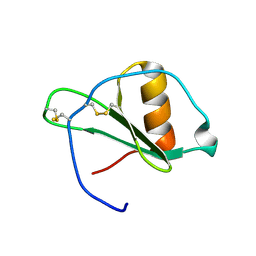 | |
1PCN
 
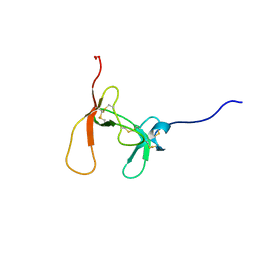 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PROCOLIPASE AS DETERMINED FROM 1H HOMONUCLEAR TWO-AND THREE-DIMENSIONAL NMR | | Descriptor: | PORCINE PANCREATIC PROCOLIPASE B | | Authors: | Breg, J.N, Sarda, L, Cozzone, P.J, Rugani, N, Boelens, R, Kaptein, R. | | Deposit date: | 1994-06-08 | | Release date: | 1994-12-20 | | Last modified: | 2024-09-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic procolipase as determined from 1H homonuclear two-dimensional and three-dimensional NMR.
Eur.J.Biochem., 227, 1995
|
|
1PCO
 
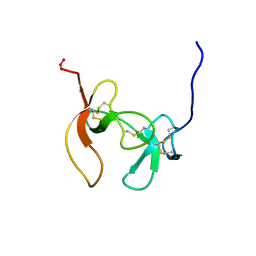 | | SOLUTION STRUCTURE OF PORCINE PANCREATIC PROCOLIPASE AS DETERMINED FROM 1H HOMONUCLEAR TWO-AND THREE-DIMENSIONAL NMR | | Descriptor: | PORCINE PANCREATIC PROCOLIPASE B | | Authors: | Breg, J.N, Sarda, L, Cozzone, P.J, Rugani, N, Boelens, R, Kaptein, R. | | Deposit date: | 1994-06-08 | | Release date: | 1994-12-20 | | Last modified: | 2024-09-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of porcine pancreatic procolipase as determined from 1H homonuclear two-dimensional and three-dimensional NMR.
Eur.J.Biochem., 227, 1995
|
|
1QLK
 
 | | SOLUTION STRUCTURE OF CA(2+)-LOADED RAT S100B (BETABETA) NMR, 20 STRUCTURES | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Drohat, A.C, Baldisseri, D.M, Rustandi, R.R, Weber, D.J. | | Deposit date: | 1997-09-26 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of calcium-bound rat S100B(betabeta) as determined by nuclear magnetic resonance spectroscopy,.
Biochemistry, 37, 1998
|
|
1EZC
 
 | |
