1DR3
 
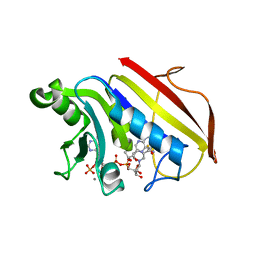 | | 2.3 ANGSTROMS CRYSTAL STRUCTURE OF CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXED WITH THIONADP+ AND BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, CALCIUM ION, ... | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of chicken liver dihydrofolate reductase: binary thioNADP+ and ternary thioNADP+.biopterin complexes.
Biochemistry, 32, 1993
|
|
1L86
 
 | |
1L90
 
 | |
1L95
 
 | |
1LGA
 
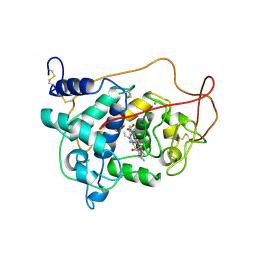 | | CRYSTALLOGRAPHIC REFINEMENT OF LIGNIN PEROXIDASE AT 2 ANGSTROMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, LIGNIN PEROXIDASE, ... | | Authors: | Poulos, T.L, Edwards, S.L, Wariishi, H, Gold, M.H. | | Deposit date: | 1992-12-08 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystallographic refinement of lignin peroxidase at 2 A.
J.Biol.Chem., 268, 1993
|
|
1DR2
 
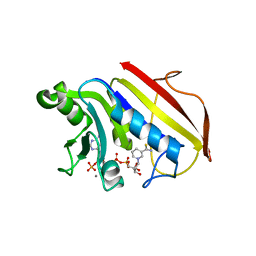 | | 2.3 ANGSTROMS CRYSTAL STRUCTURE OF CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXED WITH THIONADP+ AND BIOPTERIN | | Descriptor: | 7-THIONICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, CALCIUM ION, DIHYDROFOLATE REDUCTASE | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of chicken liver dihydrofolate reductase: binary thioNADP+ and ternary thioNADP+.biopterin complexes.
Biochemistry, 32, 1993
|
|
1DR4
 
 | | CRYSTAL STRUCTURES OF ORGANOMERCURIAL-ACTIVATED CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXES | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CALCIUM ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Organomercurial-Activated Chicken Liver Dihydrofolate Reductase Complexes
To be Published
|
|
1IFH
 
 | |
1IXA
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE FIRST EGF-LIKE MODULE OF HUMAN FACTOR IX: COMPARISON WITH EGF AND TGF-A | | Descriptor: | EGF-LIKE MODULE OF HUMAN FACTOR IX | | Authors: | Baron, M, Norman, D.G, Harvey, T.S, Hanford, P.A, Mayhew, M, Tse, A.G.D, Brownlee, G.G, Campbell, I.D.C. | | Deposit date: | 1991-11-14 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the first EGF-like module of human factor IX: comparison with EGF and TGF-alpha.
Protein Sci., 1, 1992
|
|
3FX2
 
 | |
2EDA
 
 | |
1NCP
 
 | |
2MIP
 
 | | CRYSTAL STRUCTURE OF HUMAN IMMUNODEFICIENCY VIRUS (HIV) TYPE 2 PROTEASE IN COMPLEX WITH A REDUCED AMIDE INHIBITOR AND COMPARISON WITH HIV-1 PROTEASE STRUCTURES | | Descriptor: | HIV-2 PROTEASE, INHIBITOR BI-LA-398 | | Authors: | Tong, L, Pav, S, Pargellis, C, Do, F, Lamarre, D, Anderson, P.C. | | Deposit date: | 1993-06-03 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human immunodeficiency virus (HIV) type 2 protease in complex with a reduced amide inhibitor and comparison with HIV-1 protease structures.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1NDK
 
 | | X-RAY STRUCTURE OF NUCLEOSIDE DIPHOSPHATE KINASE | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Dumas, C, Morera, S, Lascu, I, Veron, M. | | Deposit date: | 1993-07-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of nucleoside diphosphate kinase.
EMBO J., 11, 1992
|
|
1GPR
 
 | |
1DR1
 
 | | 2.2 ANGSTROMS CRYSTAL STRUCTURE OF CHICKEN LIVER DIHYDROFOLATE REDUCTASE COMPLEXED WITH NADP+ AND BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, CALCIUM ION, DIHYDROFOLATE REDUCTASE, ... | | Authors: | Mctigue, M.A, Davies /II, J.F, Kaufman, B.T, Xuong, N.-H, Kraut, J. | | Deposit date: | 1992-03-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of chicken liver dihydrofolate reductase complexed with NADP+ and biopterin.
Biochemistry, 31, 1992
|
|
1DYD
 
 | |
1NAG
 
 | | CREVICE-FORMING MUTANTS IN THE RIGID CORE OF BOVINE PANCREATIC TRYPSIN INHIBITOR: CRYSTAL STRUCTURES OF F22A, Y23A, N43G, AND F45A | | Descriptor: | BOVINE PANCREATIC TRYPSIN INHIBITOR, PHOSPHATE ION | | Authors: | Danishefsky, A.T, Wlodawer, A, Kim, K.-S, Tao, F, Woodward, C. | | Deposit date: | 1992-08-18 | | Release date: | 1993-10-31 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crevice-forming mutants in the rigid core of bovine pancreatic trypsin inhibitor: crystal structures of F22A, Y23A, N43G, and F45A.
Protein Sci., 2, 1993
|
|
1DYF
 
 | |
1NIP
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF THE NITROGENASE IRON PROTEIN FROM AZOTOBACTER VINELANDII | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Komiya, H, Georgiadis, M.M, Chakrabarti, P, Woo, D, Kornuc, J.J, Rees, D.C. | | Deposit date: | 1992-09-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii.
Science, 257, 1992
|
|
3SC2
 
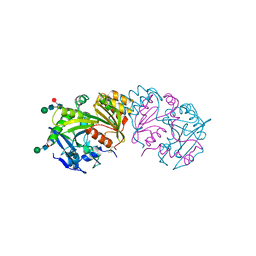 | | REFINED ATOMIC MODEL OF WHEAT SERINE CARBOXYPEPTIDASE II AT 2.2-ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SERINE CARBOXYPEPTIDASE II (CPDW-II), alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Liao, D.-I, Remington, S.J. | | Deposit date: | 1992-07-01 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined atomic model of wheat serine carboxypeptidase II at 2.2-A resolution.
Biochemistry, 31, 1992
|
|
1DYC
 
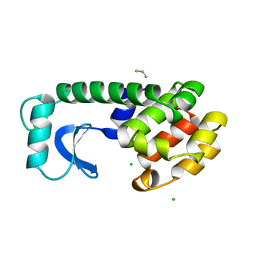 | |
1L00
 
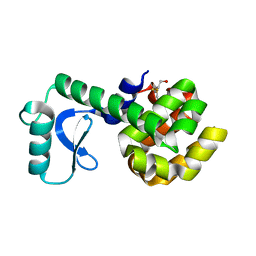 | | PERTURBATION OF TRP 138 IN T4 LYSOZYME BY MUTATIONS AT GLN 105 USED TO CORRELATE CHANGES IN STRUCTURE, STABILITY, SOLVATION, AND SPECTROSCOPIC PROPERTIES | | Descriptor: | BETA-MERCAPTOETHANOL, T4 LYSOZYME | | Authors: | Pjura, P, Mcintosh, L.P, Wozniak, J.A, Matthews, B.W. | | Deposit date: | 1992-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Perturbation of Trp 138 in T4 lysozyme by mutations at Gln 105 used to correlate changes in structure, stability, solvation, and spectroscopic properties.
Proteins, 15, 1993
|
|
4FX2
 
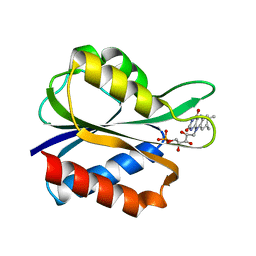 | |
1FBE
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, ZINC ION | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
