1JDT
 
 | | CRYSTAL STRUCTURE OF 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH MTA AND SULFATE ION | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, SULFATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1JDU
 
 | | CRYSTAL STRUCTURE OF 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE | | Descriptor: | 5'-METHYLTHIOADENOSINE PHOSPHORYLASE | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1JDV
 
 | | CRYSTAL STRUCTURE OF 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH ADENOSINE AND SULFATE ION | | Descriptor: | 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, ADENOSINE, SULFATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1JDW
 
 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | BETA-MERCAPTOETHANOL, L-ARGININE:GLYCINE AMIDINOTRANSFERASE | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-22 | | Release date: | 1998-01-28 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
1JDX
 
 | |
1JDY
 
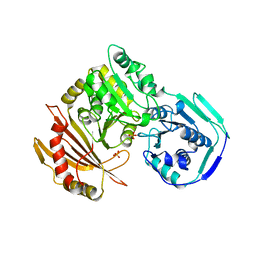 | |
1JDZ
 
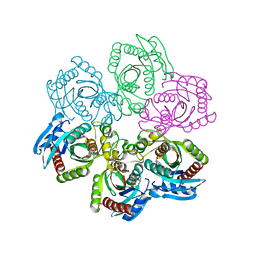 | | CRYSTAL STRUCTURE OF 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE WITH FORMYCIN B AND SULFATE ION | | Descriptor: | 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, FORMYCIN B, SULFATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1JE0
 
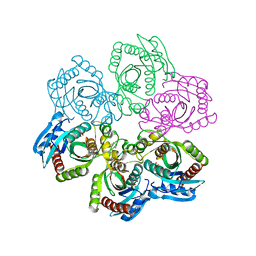 | | CRYSTAL STRUCTURE OF 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEXED WITH PHOSPHATE AND TRIS MOLECULE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, PHOSPHATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1JE1
 
 | | 5'-DEOXY-5'-METHYLTHIOADENOSINE PHOSPHORYLASE COMPLEX WITH GUANOSINE AND SULFATE | | Descriptor: | 5'-METHYLTHIOADENOSINE PHOSPHORYLASE, GUANOSINE, SULFATE ION | | Authors: | Appleby, T.C, Mathews, I.I, Porcelli, M, Cacciapuoti, G, Ealick, S.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of a hyperthermophilic 5'-deoxy-5'-methylthioadenosine phosphorylase from Sulfolobus solfataricus.
J.Biol.Chem., 276, 2001
|
|
1JE3
 
 | | Solution Structure of EC005 from Escherichia coli | | Descriptor: | HYPOTHETICAL 8.6 KDA PROTEIN IN AMYA-FLIE INTERGENIC REGION | | Authors: | Yee, A, Gutierrez, P, Kozlov, G, Denisov, A, Gehring, K, Arrowsmith, C. | | Deposit date: | 2001-06-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1JE4
 
 | |
1JE5
 
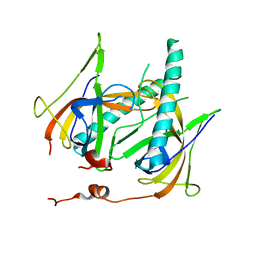 | | Crystal Structure of gp2.5, a Single-Stranded DNA Binding Protein Encoded by Bacteriophage T7 | | Descriptor: | CALCIUM ION, HELIX-DESTABILIZING PROTEIN | | Authors: | Hollis, T, Stattel, J.M, Walther, D.S, Richardson, C.C, Ellenberger, T.E. | | Deposit date: | 2001-06-15 | | Release date: | 2001-08-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JE6
 
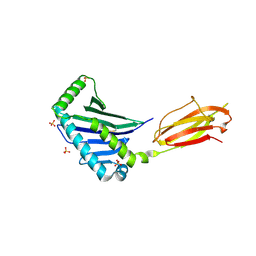 | | Structure of the MHC Class I Homolog MICB | | Descriptor: | MHC class I chain-related protein, SULFATE ION | | Authors: | Holmes, M.A, Li, P, Strong, R.K. | | Deposit date: | 2001-06-15 | | Release date: | 2002-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies of allelic diversity of the MHC class I homolog MIC-B, a stress-inducible ligand for the activating immunoreceptor NKG2D.
J.Immunol., 169, 2002
|
|
1JE8
 
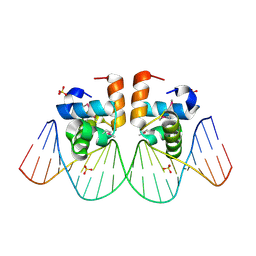 | | Two-Component response regulator NarL/DNA Complex: DNA Bending Found in a High Affinity Site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*AP*TP*TP*AP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/Nitrite Response Regulator Protein NARL, SULFATE ION | | Authors: | Maris, A.E, Sawaya, M.R, Kaczor-Grzeskowiak, M, Jarvis, M.R, Bearson, S.M.D, Kopka, M.L, Schroder, I, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2001-06-15 | | Release date: | 2002-09-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Dimerization allows DNA target site recognition by the NarL response regulator.
Nat.Struct.Biol., 9, 2002
|
|
1JE9
 
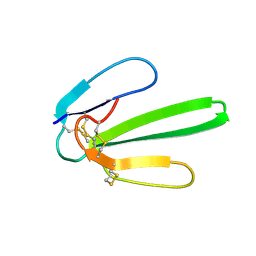 | | NMR SOLUTION STRUCTURE OF NT2 | | Descriptor: | SHORT NEUROTOXIN II | | Authors: | Cheng, Y, Wang, W, Wang, J. | | Deposit date: | 2001-06-16 | | Release date: | 2001-07-04 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of three neurotoxins from the venom of Naja kaouthia: a comparison between the NMR-derived structure of NT2 with its homologues, NT1 and NT3
BIOCHIM.BIOPHYS.ACTA, 1594, 2002
|
|
1JEA
 
 | |
1JEB
 
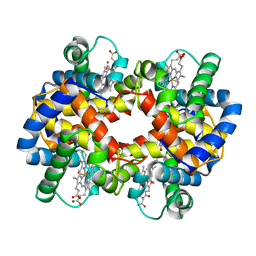 | | Chimeric Human/Mouse Carbonmonoxy Hemoglobin (Human Zeta2 / Mouse Beta2) | | Descriptor: | CARBON MONOXIDE, HEMOGLOBIN BETA-SINGLE CHAIN, HEMOGLOBIN ZETA CHAIN, ... | | Authors: | Kidd, R.D, Russell, J.E, Watmough, N.J, Baker, E.N, Brittain, T. | | Deposit date: | 2001-06-17 | | Release date: | 2002-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of beta chains in the control of the hemoglobin oxygen binding function: chimeric human/mouse proteins, structure, and function.
Biochemistry, 40, 2001
|
|
1JEC
 
 | |
1JED
 
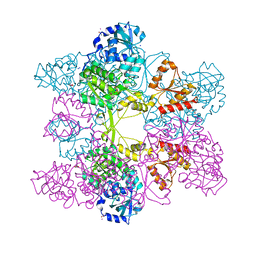 | | Crystal Structure of ATP Sulfurylase in complex with ADP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Ullrich, T.C, Huber, R. | | Deposit date: | 2001-06-17 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The complex structures of ATP sulfurylase with thiosulfate, ADP and chlorate reveal new insights in inhibitory effects and the catalytic cycle.
J.Mol.Biol., 313, 2001
|
|
1JEE
 
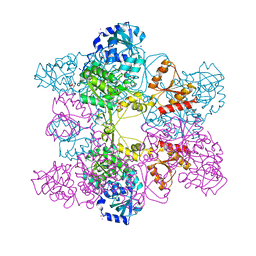 | | Crystal Structure of ATP Sulfurylase in complex with chlorate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Ullrich, T.C, Huber, R. | | Deposit date: | 2001-06-17 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The complex structures of ATP sulfurylase with thiosulfate, ADP and chlorate reveal new insights in inhibitory effects and the catalytic cycle.
J.Mol.Biol., 313, 2001
|
|
1JEF
 
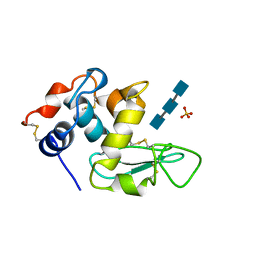 | | TURKEY LYSOZYME COMPLEX WITH (GLCNAC)3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSOZYME, SULFATE ION | | Authors: | Harata, K, Muraki, M. | | Deposit date: | 1997-04-23 | | Release date: | 1997-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | X-ray structure of turkey-egg lysozyme complex with tri-N-acetylchitotriose. Lack of binding ability at subsite A.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
1JEG
 
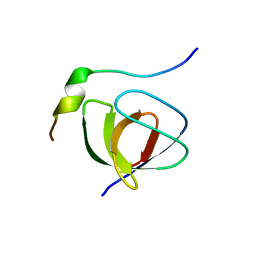 | | Solution structure of the SH3 domain from C-terminal Src Kinase complexed with a peptide from the tyrosine phosphatase PEP | | Descriptor: | HEMATOPOIETIC CELL PROTEIN-TYROSINE PHOSPHATASE 70Z-PEP, TYROSINE-PROTEIN KINASE CSK | | Authors: | Ghose, R, Shekhtman, A, Goger, M.J, Ji, H, Cowburn, D. | | Deposit date: | 2001-06-17 | | Release date: | 2001-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel, specific interaction involving the Csk SH3 domain and its natural ligand.
Nat.Struct.Biol., 8, 2001
|
|
1JEH
 
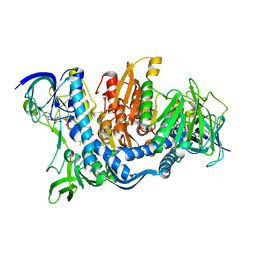 | | CRYSTAL STRUCTURE OF YEAST E3, LIPOAMIDE DEHYDROGENASE | | Descriptor: | DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Toyoda, T, Suzuki, K, Sekigushi, T, Reed, J, Takenaka, A. | | Deposit date: | 2001-06-18 | | Release date: | 2001-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of eucaryotic E3, lipoamide dehydrogenase from yeast.
J.Biochem., 123, 1998
|
|
1JEI
 
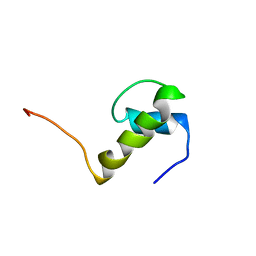 | | LEM DOMAIN OF HUMAN INNER NUCLEAR MEMBRANE PROTEIN EMERIN | | Descriptor: | EMERIN | | Authors: | Wolff, N, Gilquin, B, Courchay, K, Callebaut, I, Zinn-Justin, S. | | Deposit date: | 2001-06-18 | | Release date: | 2001-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of emerin, an inner nuclear membrane protein mutated in X-linked Emery-Dreifuss muscular dystrophy
FEBS LETT., 501, 2001
|
|
1JEJ
 
 | | T4 phage apo BGT | | Descriptor: | DNA BETA-GLUCOSYLTRANSFERASE | | Authors: | Morera, S, Lariviere, L, Kurzeck, J, Aschke-Sonnenborn, U, Freemont, P.S, Janin, J, Ruger, W. | | Deposit date: | 2001-06-18 | | Release date: | 2001-08-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High resolution crystal structures of T4 phage beta-glucosyltransferase: induced fit and effect of substrate and metal binding.
J.Mol.Biol., 311, 2001
|
|
