1H1H
 
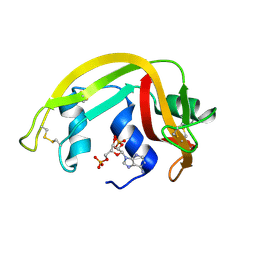 | | Crystal Structure of Eosinophil Cationic Protein in Complex with 2',5'-ADP at 2.0 A resolution Reveals the Details of the Ribonucleolytic Active site | | Descriptor: | ADENOSINE-2'-5'-DIPHOSPHATE, EOSINOPHIL CATIONIC PROTEIN | | Authors: | Mohan, C.G, Boix, E, Evans, H.R, Nikolovski, Z, Nogues, M.V, Cuchillo, C.M, Acharya, K.R. | | Deposit date: | 2002-07-15 | | Release date: | 2002-10-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Eosinophil Cationic Protein in Complex with 2'5'-Adp at 2.0 A Resolution Reveals the Details of the Ribonucleolytic Active Site
Biochemistry, 41, 2002
|
|
2QCL
 
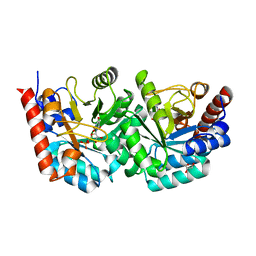 | |
5RWL
 
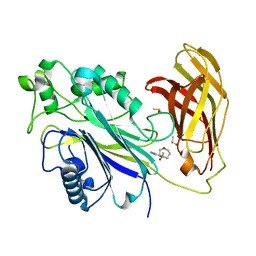 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1348371854 | | Descriptor: | 5-(1,4-oxazepan-4-yl)pyridine-2-carbonitrile, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
4QKK
 
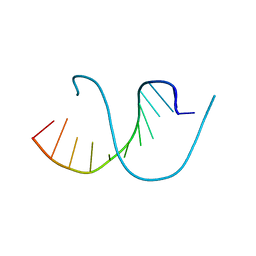 | | Crystal structure of an oligonucleotide containing 5-formylcytosine | | Descriptor: | DNA (5'-D(*CP*TP*AP*(5FC)P*GP*(5FC)P*GP*(5FC)P*GP*TP*AP*G)-3') | | Authors: | Raiber, E.-A, Murat, P, Chirgadze, D.Y, Luisi, B.F, Balasubramanian, S. | | Deposit date: | 2014-06-06 | | Release date: | 2014-12-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 5-Formylcytosine alters the structure of the DNA double helix.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5RWS
 
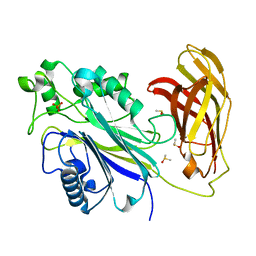 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z1478435544 | | Descriptor: | 5-methyl-N-[(pyridin-4-yl)methyl]pyridin-3-amine, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
4R88
 
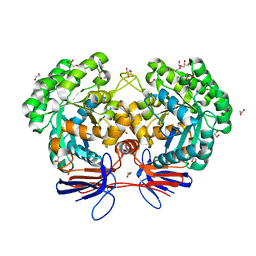 | | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-fluorocytosine | | Descriptor: | 1,2-ETHANEDIOL, 5-fluorocytosine, ACETIC ACID, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hitchcock, D.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-08-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal structure of 5-methylcytosine deaminase from Klebsiella pneumoniae liganded with 5-fluorocytosine
To be Published
|
|
5AGP
 
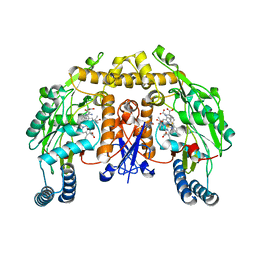 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (S)-2-Amino-5-(2-mercaptoacetimidamido)pentanoic acid | | Descriptor: | (S)-2-AMINO-5-(2-MERCAPTOACETIMIDAMIDO)PENTANOIC ACID, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
5AGO
 
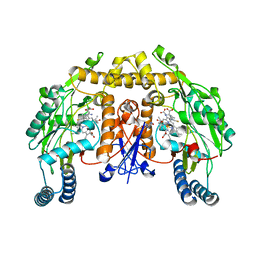 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (S)-2-Amino-5-(2-mercaptoacetimidamido)pentanoic acid | | Descriptor: | (S)-2-AMINO-5-(2-MERCAPTOACETIMIDAMIDO)PENTANOIC ACID, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
435D
 
 | | 5'-R(*UP*AP*GP*CP*CP*CP*C)-3', 5'-R(*GP*GP*GP*GP*CP*UP*A)-3' | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*CP*CP*C)-3'), SODIUM ION | | Authors: | Mueller, U, Schuebel, H, Sprinzl, M, Heinemann, U. | | Deposit date: | 1998-10-23 | | Release date: | 1999-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of acceptor stem of tRNA(Ala) from Escherichia coli shows unique G.U wobble base pair at 1.16 A resolution.
RNA, 5, 1999
|
|
3LRR
 
 | | Crystal structure of human RIG-I CTD bound to a 12 bp AU rich 5' ppp dsRNA | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*(ATP)P*UP*AP*UP*AP*UP*AP*UP*AP*UP*AP*U)-3'), ZINC ION | | Authors: | Li, P. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structural Basis of 5' Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain.
Structure, 18, 2010
|
|
1QJ6
 
 | | Novel Covalent Active Site Thrombin Inhibitors | | Descriptor: | 6-CARBAMIMIDOYL-2-[2-HYDROXY-5-(3-METHOXY-PHENYL)-INDAN-1-YL]-HEXANOIC ACID, HIRUGEN, THROMBIN | | Authors: | Jhoti, H, Cleasby, A. | | Deposit date: | 1999-06-22 | | Release date: | 2000-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Thrombin Complexed to a Novel Series of Synthetic Inhibitors Containing a 5,5-Trans-Lactone Template
Biochemistry, 38, 1999
|
|
5AGL
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (S)-2-Amino-5-(2-(methylsulfonyl)acetimidamido)pentanoic acid | | Descriptor: | (S)-2-Amino-5-(2-(methylsulfonyl)acetimidamido)pentanoic acid, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
3UPB
 
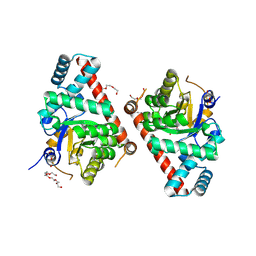 | | 1.5 Angstrom Resolution Crystal Structure of Transaldolase from Francisella tularensis in Covalent Complex with Arabinose-5-Phosphate | | Descriptor: | ARABINOSE-5-PHOSPHATE, DI(HYDROXYETHYL)ETHER, HEXAETHYLENE GLYCOL, ... | | Authors: | Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-17 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Arabinose 5-phosphate covalently inhibits transaldolase.
J.Struct.Funct.Genom., 15, 2014
|
|
7D0Z
 
 | |
2XZF
 
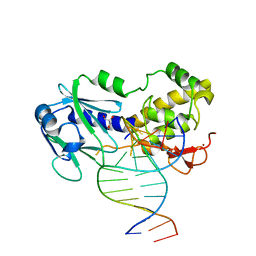 | | CRYSTAL STRUCTURE OF A COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND AN OXIDIZED PYRIMIDINE CONTAINING DNA AT 293K | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*VETP*TP*TP*TP*CP*TP*CP*GP)-3', 5'-D(*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*AP*GP*CP)-3', FORMAMIDOPYRIMIDINE-DNA GLYCOSYLASE, ... | | Authors: | LeBihan, Y.V, Izquierdo, M.A, Coste, F, Culard, F, Gehrke, T.H, Essalhi, K, Aller, P, Carrel, T, Castaing, B. | | Deposit date: | 2010-11-25 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | 5-Hydroxy-5-Methylhydantoin DNA Lesion, a Molecular Trap for DNA Glycosylases
Nucleic Acids Res., 39, 2011
|
|
3LRN
 
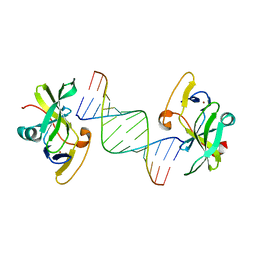 | | Crystal structure of human RIG-I CTD bound to a 14 bp GC 5' ppp dsRNA | | Descriptor: | Probable ATP-dependent RNA helicase DDX58, RNA (5'-R(*(GTP)P*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ZINC ION | | Authors: | Li, P. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of 5' Triphosphate Double-Stranded RNA Recognition by RIG-I C-Terminal Domain.
Structure, 18, 2010
|
|
5AGM
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (S)-2-Amino-5-(2-oxoacetimidamido)pentanoic acid | | Descriptor: | (S)-2-AMINO-5-(2-OXOACETIMIDAMIDO)PENTANOIC ACID, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-02-02 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Mechanism of Inactivation of Neuronal Nitric Oxide Synthase by (S)-2-Amino-5-(2-(Methylthio)Acetimidamido)Pentanoic Acid.
J.Am.Chem.Soc., 137, 2015
|
|
434D
 
 | | 5'-R(*UP*AP*GP*CP*UP*CP*C)-3', 5'-R(*GP*GP*GP*GP*CP*UP*A)-3' | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*CP*UP*A)-3'), RNA (5'-R(*UP*AP*GP*CP*UP*CP*C)-3'), SODIUM ION, ... | | Authors: | Mueller, U, Schuebel, H, Sprinzl, M, Heinemann, U. | | Deposit date: | 1998-10-23 | | Release date: | 1999-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of acceptor stem of tRNA(Ala) from Escherichia coli shows unique G.U wobble base pair at 1.16 A resolution.
RNA, 5, 1999
|
|
4FCR
 
 | | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization | | Descriptor: | 2-{[4-(2-chloro-4,5-dimethoxyphenyl)-5-cyano-7H-pyrrolo[2,3-d]pyrimidin-2-yl]sulfanyl}-N,N-dimethylacetamide, Heat shock protein HSP 90-alpha | | Authors: | Davies, N.G, Browne, H, Davis, B, Foloppe, N, Geoffrey, S, Gibbons, B, Hart, T, Drysdale, M.J, Mansell, H, Massey, A, Matassova, N, Moore, J.D, Murray, J, Pratt, R, Ray, S, Roughley, S.D, Jensen, M.R, Schoepfer, J, Scriven, K, Simmonite, H, Stokes, S, Surgenor, A, Webb, P, Wright, L, Brough, P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization.
Bioorg.Med.Chem., 20, 2012
|
|
9JHV
 
 | | Crystal Structure of 5'-Deoxy-5'-methylthioadenosine phosphorylase from Aeropyrum pernix complex with 5'-Deoxy-5'-methylthioadenosine 343K | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Iizuka, Y, Kikuchi, M, Yamauchi, T, Tsunoda, M. | | Deposit date: | 2024-09-10 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of 5'-Deoxy-5'-methylthioadenosine phosphorylase from Aeropyrum pernix complex with 5'-Deoxy-5'-methylthioadenosine 353K
To Be Published
|
|
3L0K
 
 | |
2FJP
 
 | | Human dipeptidyl peptidase IV/CD26 in complex with an inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(4-{(1S,2S)-2-AMINO-1-[(DIMETHYLAMINO)CARBONYL]-3-[(3S)-3-FLUOROPYRROLIDIN-1-YL]-3-OXOPROPYL}PHENYL)-1H-[1,2,4]TRIAZOLO[1,5-A]PYRIDIN-4-IUM, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W. | | Deposit date: | 2006-01-03 | | Release date: | 2006-07-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | (2S,3S)-3-Amino-4-(3,3-difluoropyrrolidin-1-yl)-N,N-dimethyl-4-oxo-2-(4-[1,2,4]triazolo[1,5-a]- pyridin-6-ylphenyl)butanamide: a selective alpha-amino amide dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes.
J.Med.Chem., 49, 2006
|
|
9JD2
 
 | |
1JBQ
 
 | | STRUCTURE OF HUMAN CYSTATHIONINE BETA-SYNTHASE: A UNIQUE PYRIDOXAL 5'-PHOSPHATE DEPENDENT HEMEPROTEIN | | Descriptor: | CYSTATHIONINE BETA-SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Meier, M, Janosik, M, Kery, V, Kraus, J.P, Burkhard, P. | | Deposit date: | 2001-06-06 | | Release date: | 2001-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human cystathionine beta-synthase: a unique pyridoxal 5'-phosphate-dependent heme protein.
EMBO J., 20, 2001
|
|
1FPG
 
 | | STRUCTURAL ASPECTS OF THE ALLOSTERIC INHIBITION OF FRUCTOSE-1,6-BISPHOSPHATASE BY AMP: THE BINDING OF BOTH THE SUBSTRATE ANALOGUE 2,5-ANHYDRO-D-GLUCITOL-1,6-BISPHOSPHATE AND CATALYTIC METAL IONS MONITORED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | Authors: | Villeret, V, Huang, S, Zhang, Y, Lipscomb, W.N. | | Deposit date: | 1994-12-15 | | Release date: | 1995-02-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural aspects of the allosteric inhibition of fructose-1,6-bisphosphatase by AMP: the binding of both the substrate analogue 2,5-anhydro-D-glucitol 1,6-bisphosphate and catalytic metal ions monitored by X-ray crystallography.
Biochemistry, 34, 1995
|
|
