1VH9
 
 | | Crystal structure of a putative thioesterase | | Descriptor: | Hypothetical protein ybdB | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
5YUN
 
 | | Crystal structure of SSB complexed with myc | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Single-stranded DNA-binding protein | | Authors: | Huang, Y.H, Huang, C.Y. | | Deposit date: | 2017-11-22 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of SSB complexed with inhibitor myricetin.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
6A6K
 
 | | Crystal structure of Estrogen-related Receptor-3 (ERR-gamma) ligand binding domain with DN201000 | | Descriptor: | 3-[(~{E})-5-oxidanyl-2-phenyl-1-[4-(4-propan-2-ylpiperazin-1-yl)phenyl]pent-1-enyl]phenol, Estrogen-related receptor gamma | | Authors: | Yoon, H, Kim, J, Chin, J, Cho, S.J, Song, J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Estrogen-Related Receptor-gamma Inverse Agonists To Restore the Sodium Iodide Symporter Function in Anaplastic Thyroid Cancer.
J. Med. Chem., 62, 2019
|
|
1VH4
 
 | |
1VHO
 
 | |
1VHY
 
 | |
1VIY
 
 | | Crystal structure of dephospho-CoA kinase | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
6A83
 
 | |
1VH7
 
 | |
1VHA
 
 | |
1VHF
 
 | |
1VHZ
 
 | | Crystal structure of ADP compounds hydrolase | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP compounds hydrolase nudE | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHU
 
 | | Crystal structure of a putative phosphoesterase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hypothetical protein AF1521 | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VI5
 
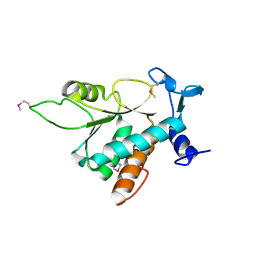 | | Crystal structure of ribosomal protein S2P | | Descriptor: | 30S ribosomal protein S2P | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIS
 
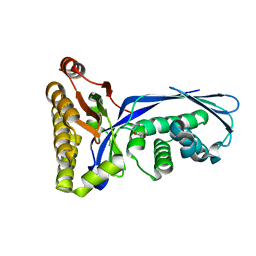 | | Crystal structure of mevalonate kinase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Mevalonate kinase | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
6AH6
 
 | | M500V mutant of Coronin coiled coil domain | | Descriptor: | Coronin-like protein | | Authors: | Ansari, A, Karade, S.S, Pratap, J.V. | | Deposit date: | 2018-08-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular and structural analysis of a mechanical transition of helices in the L. donovani coronin coiled-coil domain.
Int.J.Biol.Macromol., 143, 2020
|
|
6SLH
 
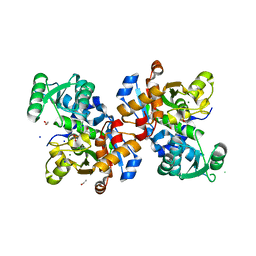 | | Conformational flexibility within the small domain of human serine racemase. | | Descriptor: | MAGNESIUM ION, SODIUM ION, Serine racemase, ... | | Authors: | Koulouris, C.R, Bax, B, Atack, J, Roe, S.M. | | Deposit date: | 2019-08-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Conformational flexibility within the small domain of human serine racemase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6ADO
 
 | | LdCoroCC mutant-I486A | | Descriptor: | Coronin-like protein | | Authors: | Karade, S.S, Ansari, A, Pratap, J.V. | | Deposit date: | 2018-08-01 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Molecular and structural analysis of a mechanical transition of helices in the L. donovani coronin coiled-coil domain.
Int.J.Biol.Macromol., 143, 2020
|
|
6A82
 
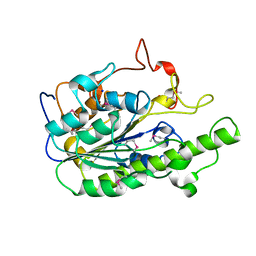 | |
6ADZ
 
 | | LdCoroCC Double mutant- I486A-L493A | | Descriptor: | Coronin-like protein, SULFATE ION | | Authors: | Karade, S.S, Ansari, A, Pratap, J.V. | | Deposit date: | 2018-08-02 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | Molecular and structural analysis of a mechanical transition of helices in the L. donovani coronin coiled-coil domain.
Int.J.Biol.Macromol., 143, 2020
|
|
5O2F
 
 | | Crystal structure of NDM-1 in complex with hydrolyzed ampicillin - new refinement | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Raczynska, J.E, Shabalin, I.G, Jaskolski, M, Minor, W, Wlodawer, A. | | Deposit date: | 2017-05-20 | | Release date: | 2018-12-26 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A close look onto structural models and primary ligands of metallo-beta-lactamases.
Drug Resist. Updat., 40, 2018
|
|
1VI2
 
 | | Crystal structure of shikimate-5-dehydrogenase with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, Shikimate 5-dehydrogenase 2 | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VIM
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | FORMIC ACID, Hypothetical protein AF1796 | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VGW
 
 | |
1VHV
 
 | | Crystal structure of diphthine synthase | | Descriptor: | diphthine synthase | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
