8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
7B3O
 
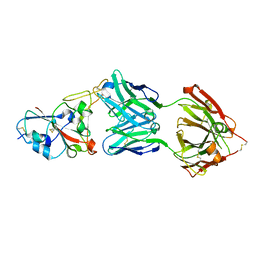 | | Crystal structure of the SARS-CoV-2 RBD in complex with STE90-C11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Fab Fragment, Light Chain of Fab Fragment, ... | | Authors: | Kluenemann, T, Van den Heuvel, J. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody selected from COVID-19 patients binds to the ACE2-RBD interface and is tolerant to most known RBD mutations.
Cell Rep, 36, 2021
|
|
8V87
 
 | | 60S ribosome biogenesis intermediate (Dbp10 post-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V83
 
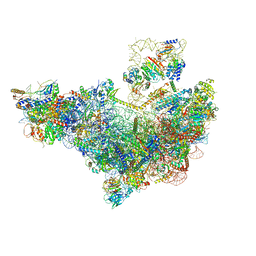 | | 60S ribosome biogenesis intermediate (Dbp10 pre-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
7DOK
 
 | |
4X1T
 
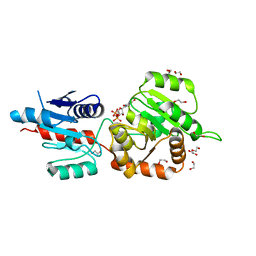 | | The crystal structure of Arabidopsis thaliana galactolipid synthase MGD1 in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, Monogalactosyldiacylglycerol synthase 1, chloroplastic, ... | | Authors: | Rocha, J, Breton, C. | | Deposit date: | 2014-11-25 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights and membrane binding properties of MGD1, the major galactolipid synthase in plants.
Plant J., 85, 2016
|
|
6LH6
 
 | |
6YLY
 
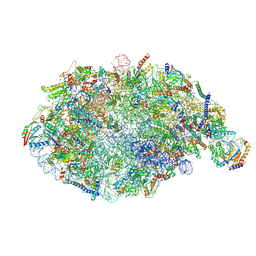 | | pre-60S State NE2 (TAP-Flag-Nop53) | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Beckmann, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Construction of the Central Protuberance and L1 Stalk during 60S Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
6YLX
 
 | | pre-60S State NE1 (TAP-Flag-Nop53) | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Kater, L, Beckmann, R. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Construction of the Central Protuberance and L1 Stalk during 60S Subunit Biogenesis.
Mol.Cell, 79, 2020
|
|
7U0H
 
 | | State NE1 nucleolar 60S ribosome biogenesis intermediate - Overall model | | Descriptor: | 25S rRNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, 5.8S rRNA, ... | | Authors: | Cruz, V.E, Sekulski, K, Peddada, N, Erzberger, J.P. | | Deposit date: | 2022-02-18 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Sequence-specific remodeling of a topologically complex RNP substrate by Spb4.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UQB
 
 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a SPB1-D52A strain with AlF4 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
7UOO
 
 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-13 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
7UQZ
 
 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a SPB1 D52A strain | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
7V08
 
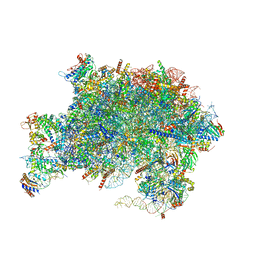 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a Spb1 D52A suppressor 3 strain | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
7SG5
 
 | | Structure of PfCSP peptide 21 with antibody CIS43_Var2 | | Descriptor: | CIS43_Var2 Fab Heavy chain, CIS43_Var2 Fab Light chain, GLYCEROL, ... | | Authors: | Tripathi, P, Kwong, P.D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Highly protective antimalarial antibodies via precision library generation and yeast display screening.
J.Exp.Med., 219, 2022
|
|
7SG6
 
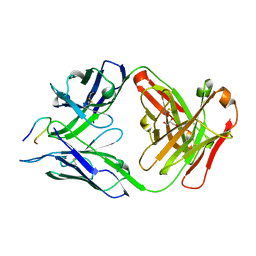 | | Structure of PfCSP peptide 21 with antibody CIS43_Var10 | | Descriptor: | CIS43_Var10 Fab Heavy chain, CIS43_Var10 Fab Light chain, GLYCEROL, ... | | Authors: | Tripathi, P, Kwong, P.D. | | Deposit date: | 2021-10-05 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Highly protective antimalarial antibodies via precision library generation and yeast display screening.
J.Exp.Med., 219, 2022
|
|
7RWN
 
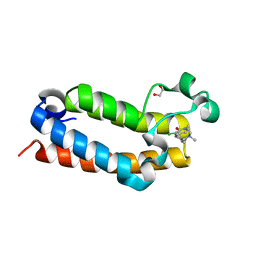 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWP
 
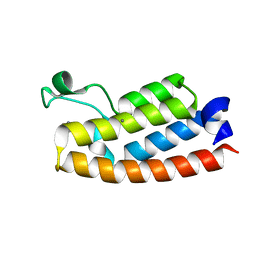 | | Crystal Structure of BPTF bromodomain in complex with 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one | | Descriptor: | 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWQ
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWO
 
 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
4EY8
 
 | | Crystal structure of recombinant human acetylcholinesterase in complex with fasciculin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, Fasciculin-2, ... | | Authors: | Cheung, J, Rudolph, M, Burshteyn, F, Cassidy, M, Gary, E, Love, J, Height, J, Franklin, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5958 Å) | | Cite: | Structures of human acetylcholinesterase in complex with pharmacologically important ligands.
J.Med.Chem., 55, 2012
|
|
5Z3G
 
 | | Cryo-EM structure of a nucleolar pre-60S ribosome (Rpf1-TAP) | | Descriptor: | 25S rRNA, 5.8S rRNA, 60S ribosomal protein L13-A, ... | | Authors: | Zhu, X, Zhou, D, Ye, K. | | Deposit date: | 2018-01-06 | | Release date: | 2018-04-11 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate.
Protein Cell, 10, 2019
|
|
6EM1
 
 | | State C (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
4U1E
 
 | | Crystal structure of the eIF3b-CTD/eIF3i/eIF3g-NTD translation initiation complex | | Descriptor: | Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit G, Eukaryotic translation initiation factor 3 subunit I | | Authors: | Zhang, S, Erzberger, J.P, Schaefer, T, Ban, N. | | Deposit date: | 2014-07-15 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Architecture of the 40SeIF1eIF3 Translation Initiation Complex.
Cell, 158, 2014
|
|
6EM3
 
 | | State A architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
