2PPX
 
 | | Crystal structure of a HTH XRE-family like protein from Agrobacterium tumefaciens | | Descriptor: | GLYCEROL, SULFATE ION, Uncharacterized protein Atu1735 | | Authors: | Cuff, M.E, Skarina, T, Onopriyenko, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-04-30 | | Release date: | 2007-05-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a HTH XRE-family like protein from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
2R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-13 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
2EBY
 
 | | Crystal structure of a hypothetical protein from E. Coli | | Descriptor: | Putative HTH-type transcriptional regulator ybaQ, SULFATE ION | | Authors: | Karthe, P, Kumarevel, T.S, Ebihara, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-09 | | Release date: | 2007-08-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of a hypothetical protein from E. Coli
To be Published
|
|
2EF8
 
 | |
1R69
 
 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | Deposit date: | 1988-12-08 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
2CRO
 
 | |
2EWT
 
 | | Crystal structure of the DNA-binding domain of BldD | | Descriptor: | SULFATE ION, putative DNA-binding protein | | Authors: | Kim, I.K, Lee, C.J, Kim, M.K, Kim, J.M, Kim, J.H, Yim, H.S, Cha, S.S, Kang, S.O. | | Deposit date: | 2005-11-07 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the DNA-binding domain of BldD, a central regulator of aerial mycelium formation in Streptomyces coelicolor A3(2)
Mol.Microbiol., 60, 2006
|
|
1SQ8
 
 | |
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
2GZU
 
 | |
1PRA
 
 | |
1RPE
 
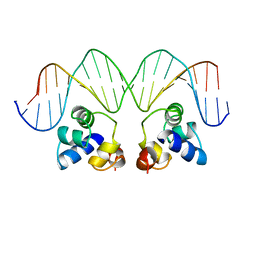 | | THE PHAGE 434 OR2/R1-69 COMPLEX AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*AP*GP*AP*TP*AP*CP*AP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*TP*GP*TP*AP*TP*CP*TP*TP*GP*T P*TP*TP*G)-3'), PROTEIN (434 REPRESSOR) | | Authors: | Shimon, L.J.W, Harrison, S.C. | | Deposit date: | 1993-03-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The phage 434 OR2/R1-69 complex at 2.5 A resolution.
J.Mol.Biol., 232, 1993
|
|
1PER
 
 | |
6IRP
 
 | |
6JPI
 
 | |
6JQ4
 
 | | HIGA Escherichia coli-K12 | | Descriptor: | Antitoxin HigA | | Authors: | She, Z, Xu, B.S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes of antitoxin HigA from Escherichia coli str. K-12 upon binding of its cognate toxin HigB reveal a new regulation mechanism in toxin-antitoxin systems.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
6JQ1
 
 | | Crystal Structure of DdrO from Deinococcus geothermalis | | Descriptor: | LITHIUM ION, Transcriptional regulator, XRE family | | Authors: | Lu, H, Hua, Y, Zhao, Y. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and DNA damage-dependent derepression mechanism for the XRE family member DG-DdrO.
Nucleic Acids Res., 47, 2019
|
|
4I6T
 
 | |
4I6U
 
 | | Crystal Structure of a Y37F mutant of the Restriction-Modification Controller Protein C.Esp1396I | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2012-11-30 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Mutagenic Analysis of the RM Controller Protein C.Esp1396I.
Plos One, 9, 2014
|
|
4IVZ
 
 | |
4I8T
 
 | | C.Esp1396I bound to a 19 base pair DNA duplex | | Descriptor: | DNA (5'-D(P*TP*GP*TP*GP*TP*GP*AP*TP*TP*AP*TP*AP*GP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(P*TP*GP*TP*TP*GP*AP*CP*TP*AP*TP*AP*AP*TP*CP*AP*CP*AP*CP*A)-3'), Regulatory protein | | Authors: | Martin, R.N.A, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2012-12-04 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of DNA-protein complexes regulating the restriction-modification system Esp1396I.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4JCY
 
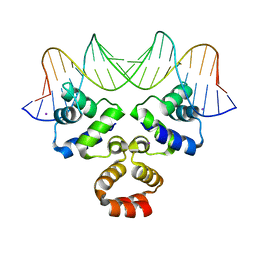 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OR operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*AP*AP*CP*TP*AP*AP*GP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*TP*AP*AP*GP*AP*TP*TP*TP*TP*CP*TP*TP*AP*GP*TP*TP*T)-3'), ... | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, Mcgeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4IWR
 
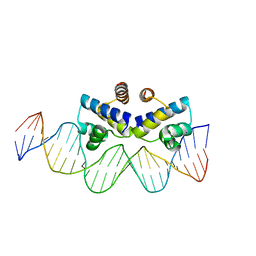 | | C.Esp1396I bound to a 25 base pair operator site | | Descriptor: | DNA (25-MER), Regulatory protein | | Authors: | Martin, R.N.A, McGeehan, J.E, Ball, N.J, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | Deposit date: | 2013-01-24 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of DNA-protein complexes regulating the restriction-modification system Esp1396I.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4JCX
 
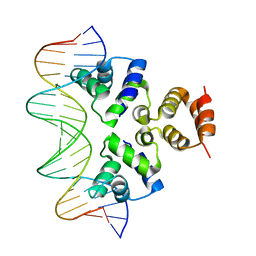 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OL operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*CP*AP*CP*TP*AP*AP*GP*GP*AP*AP*AP*AP*CP*TP*TP*AP*GP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*AP*AP*GP*TP*TP*TP*TP*CP*CP*TP*TP*AP*GP*TP*GP*T)-3') | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, Mcgeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-02-22 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4JQD
 
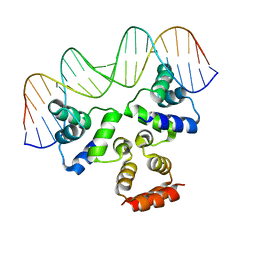 | | Crystal structure of the Restriction-Modification Controller Protein C.Csp231I OL operator complex | | Descriptor: | Csp231I C protein, DNA (5'-D(*AP*CP*AP*CP*TP*AP*AP*GP*GP*AP*AP*AP*AP*CP*TP*TP*AP*GP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*AP*AP*GP*TP*TP*TP*TP*CP*CP*TP*TP*AP*GP*TP*GP*T)-3') | | Authors: | Shevtsov, M.B, Streeter, S.D, Thresh, S.J, McGeehan, J.E, Kneale, G.G. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of DNA binding by C.Csp231I, a member of a novel class of R-M controller proteins regulating gene expression.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
