5HFG
 
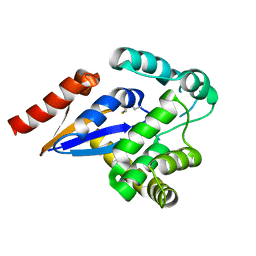 | | Cytosolic disulfide reductase DsbM from Pseudomonas aeruginosa | | Descriptor: | Uncharacterized protein, cytosolic disulfide reductase DsbM | | Authors: | Jo, I, Ha, N.-C. | | Deposit date: | 2016-01-07 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Crystal structures of the disulfide reductase DsbM from Pseudomonas aeruginosa
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5QME
 
 | |
7L76
 
 | |
5QO0
 
 | |
5QMK
 
 | |
7LUH
 
 | |
5QO1
 
 | |
8D11
 
 | |
2IN3
 
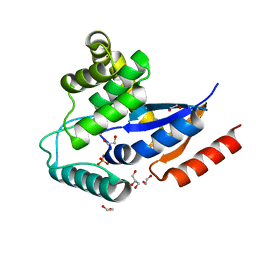 | | Crystal structure of a putative protein disulfide isomerase from Nitrosomonas europaea | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Hypothetical protein, ... | | Authors: | Cuff, M.E, Skarina, T, Onopriyenko, O, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-05 | | Release date: | 2006-11-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a putative protein disulfide isomerase from Nitrosomonas europaea
To be Published
|
|
5QNU
 
 | |
5QNJ
 
 | |
5QNM
 
 | |
5QLA
 
 | |
5QLQ
 
 | |
5QN8
 
 | |
4JRR
 
 | | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila | | Descriptor: | GLYCEROL, SULFATE ION, Thiol:disulfide interchange protein DsbA | | Authors: | Shumilin, I.A, Jameson-Lee, M, Cymborowski, M, Domagalski, M.J, Chertihin, O, Kpadeh, Z.Z, Yeh, A.J, Hoffman, P.S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-21 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of disulfide bond oxidoreductase DsbA1 from Legionella pneumophila
To be Published
|
|
5QKP
 
 | |
5QNW
 
 | |
5QLH
 
 | |
5QMC
 
 | |
5QN5
 
 | |
5QKT
 
 | |
5QKN
 
 | |
5QLX
 
 | |
5QNG
 
 | |
