7QXO
 
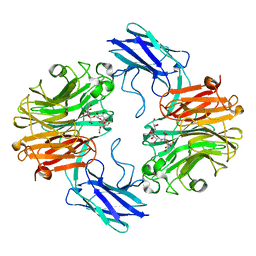 | | The structure of T. forsythia NanH | | Descriptor: | (2~{R},4~{S},5~{S},6~{S})-5-acetamido-4-oxidanyl-6-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]-2-[[(2~{R},3~{R},4~{S},5~{R},6~{S})-3,4,5-tris(oxidanyl)-6-[(2~{R},3~{S},4~{R},5~{R})-1,2,4,5-tetrakis(oxidanyl)-6-oxidanylidene-hexan-3-yl]oxy-oxan-2-yl]methoxy]oxane-2-carboxylic acid, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-26 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7QDL
 
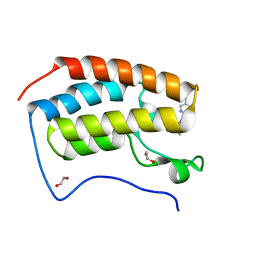 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 with I-BET567 | | Descriptor: | (2S,4R)-1-acetyl-4-((5-chloropyrimidin-2-yl)amino)-2-methyl-1,2,3,4-tetrahydroquinoline-6-carboxamide, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2021-11-27 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Design, Synthesis, and Characterization of I-BET567, a Pan-Bromodomain and Extra Terminal (BET) Bromodomain Oral Candidate.
J.Med.Chem., 65, 2022
|
|
7QEC
 
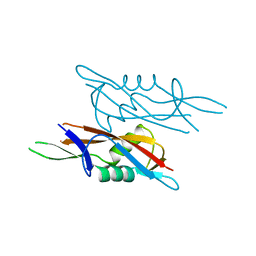 | | Crystal structure of SlpA - domain II, domain that is involved in the self-assembly of the S-layer from Lactobacillus amylovorus | | Descriptor: | S-layer | | Authors: | Eder, M, Dordic, A, Millan, C, Sagmeister, T, Uson, I, Pavkov-Keller, T. | | Deposit date: | 2021-12-02 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QY8
 
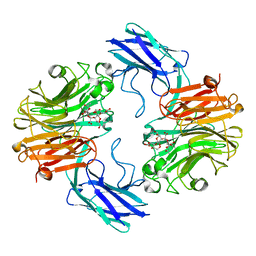 | | The structure of T. forsythia NanH | | Descriptor: | 3-sialyllactose, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7QEH
 
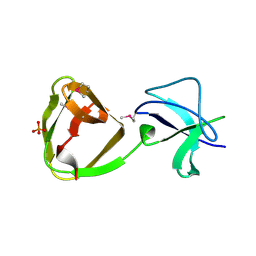 | | LTA-binding domain of SlpA, the S-layer protein from Lactobacillus amylovorus | | Descriptor: | PHOSPHATE ION, S-layer | | Authors: | Eder, M, Dordic, A, Sagmeister, T, Pavkov-Keller, T. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QY9
 
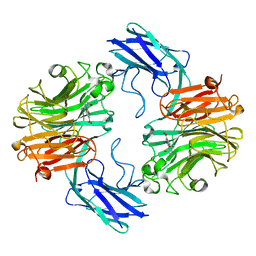 | | The structure of T.forsythia NanH with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-27 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7QZ3
 
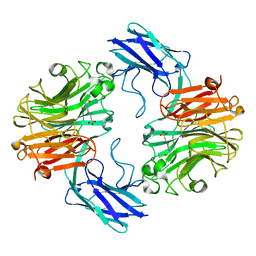 | | The structure of T. forsythia NanH | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7QYJ
 
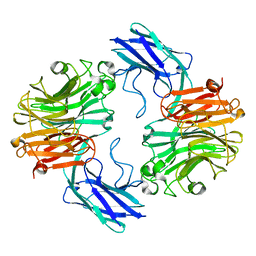 | | The structure of T. forsythia NanH | | Descriptor: | BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7QFI
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain I (aa 31-182) | | Descriptor: | CALCIUM ION, SlpX | | Authors: | Sagmeister, T, Damisch, E, Millan, C, Uson, I, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFG
 
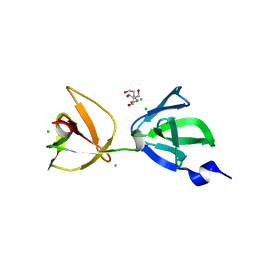 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain III (aa 309-444) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, S-layer protein | | Authors: | Sagmeister, T, Eder, M, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFK
 
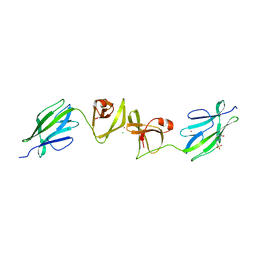 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II, Co-Crystallization with HgCl2, Mutation Ser316Cys (aa 194-362) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QHE
 
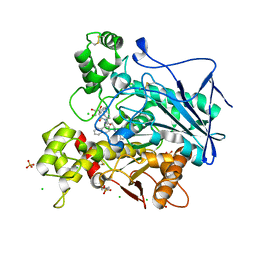 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((naphthalen-1-yl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-(naphthalen-1-ylcarbamoyl)phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
7QYP
 
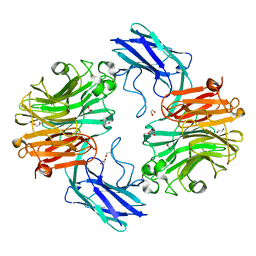 | | The structure of T. forsythia NanH | | Descriptor: | 1,2-ETHANEDIOL, BNR/Asp-box repeat protein | | Authors: | Rafferty, J, Stafford, G, Satur, M. | | Deposit date: | 2022-01-28 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Structural and functional characterisation of a stable, broad-specificity multimeric sialidase from the oral pathogen Tannerella forsythia.
Biochem.J., 479, 2022
|
|
7QFL
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain II (aa 199-308) | | Descriptor: | ACETATE ION, PHOSPHATE ION, S-layer protein | | Authors: | Sagmeister, T, Dordic, A, Eder, E, Pavkov-Keller, T. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QLH
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus amylovorus, domain I (aa 48-213) | | Descriptor: | PHOSPHATE ION, S-layer, SODIUM ION | | Authors: | Grininger, C, Sagmeister, T, Eder, E, Vejzovic, D, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QFJ
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II (aa 194-362) | | Descriptor: | SlpX | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C, Baek, M, Read, R, Baker, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QHD
 
 | | Human Butyrylcholinesterase in complex with (S)-1-(4-((2-(1H-indol-3-yl)ethyl)carbamoyl)benzyl)-N-(3-((1,2,3,4-tetrahydroacridin-9-yl)amino)propyl)piperidine-3-carboxamide | | Descriptor: | (3~{S})-1-[[4-[2-(1~{H}-indol-3-yl)ethylcarbamoyl]phenyl]methyl]-~{N}-[3-(1,2,3,4-tetrahydroacridin-9-ylamino)propyl]piperidine-3-carboxamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Jing, L, Zhan, P, Liu, X, Nachon, F. | | Deposit date: | 2021-12-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Rapid discovery and crystallography study of highly potent and selective butylcholinesterase inhibitors based on oxime-containing libraries and conformational restriction strategies.
Bioorg.Chem., 134, 2023
|
|
7QJL
 
 | | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | ACETATE ION, BROMIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Snee, M, Levy, C, Katariya, M. | | Deposit date: | 2021-12-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7QLE
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain I (aa 32-198) | | Descriptor: | S-layer protein | | Authors: | Sagmeister, T, Eder, M, Vejzovic, D, Dordic, A, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QLD
 
 | | Crystal structure of S-layer protein SlpA from Lactobacillus acidophilus, domain I, Co-crystallization with HgCl2, Mutation Ser146Cys, (aa 32-198) | | Descriptor: | CHLORIDE ION, MERCURY (II) ION, S-layer protein | | Authors: | Sagmeister, T, Vejzovic, D, Eder, M, Dordic, A, Pavkov-Keller, T. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7QKE
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with inhibitor (surface entropy reduction mutant) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid C26-monooxygenase, ethyl 1-(cyclohexylmethyl)-5-pyridin-4-yl-indole-2-carboxylate | | Authors: | Snee, M, Tunnicliffe, R, Leys, D, Levy, C, Katariya, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7QNN
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with inhibitor (surface entropy reduction mutant) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid C26-monooxygenase, ethyl 1-(cyclopentylmethyl)-5-pyridin-4-yl-indole-2-carboxylate | | Authors: | Snee, M, Tunnicliffe, R, Leys, D, Levy, C, Katariya, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7QRA
 
 | | Crystal structure of CK1 delta in complex with VN725 | | Descriptor: | 1,2-ETHANEDIOL, 4-[3-cyclohexyl-5-(4-fluorophenyl)imidazol-4-yl]-1~{H}-pyrrolo[2,3-b]pyridine, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Nemec, V, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7QR9
 
 | | Crystal structure of CK1 delta in complex with PK-09-82 | | Descriptor: | 1,2-ETHANEDIOL, 4-[5-(4-fluorophenyl)-3-(pyridin-4-ylmethyl)imidazol-4-yl]-1~{H}-pyrrolo[2,3-b]pyridine, Casein kinase I isoform delta, ... | | Authors: | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7QRB
 
 | | Crystal structure of CK1 delta in complex with PK-09-129 | | Descriptor: | 3-(dimethylamino)-~{N}-[4-[4-(4-fluorophenyl)-5-(1~{H}-pyrrolo[2,3-b]pyridin-4-yl)imidazol-1-yl]cyclohexyl]propane-1-sulfonamide, Casein kinase I isoform delta, SULFATE ION | | Authors: | Chaikuad, A, Khirsariya, P, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-01-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Potent and Exquisitely Selective Inhibitors of Kinase CK1 with Tunable Isoform Selectivity.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
