1BA8
 
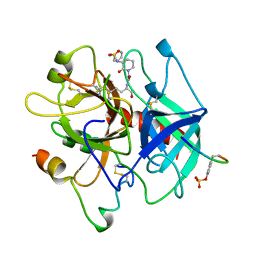 | | THROMBIN INHIBITOR WITH A RIGID TRIPEPTIDYL ALDEHYDES | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIRUGEN, THROMBIN, ... | | Authors: | Krishnan, R, Zhang, E, Hakansson, K, Arni, R.K, Tulinsky, A, Lim-Wilby, M.S.L, Levy, O.E, Semple, J.E, Brunck, T.K. | | Deposit date: | 1998-04-23 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes.
Biochemistry, 37, 1998
|
|
1BBW
 
 | | LYSYL-TRNA SYNTHETASE (LYSS) | | Descriptor: | PROTEIN (LYSYL-TRNA SYNTHETASE) | | Authors: | Onesti, S, Desogus, G, Brevet, A, Chen, J, Plateau, P, Blanquet, S, Brick, P. | | Deposit date: | 1998-04-24 | | Release date: | 2000-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding.
Biochemistry, 39, 2000
|
|
1BA9
 
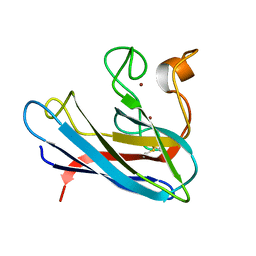 | | THE SOLUTION STRUCTURE OF REDUCED MONOMERIC SUPEROXIDE DISMUTASE, NMR, 36 STRUCTURES | | Descriptor: | COPPER (I) ION, SUPEROXIDE DISMUTASE, ZINC ION | | Authors: | Banci, L, Benedetto, M, Bertini, I, Del Conte, R, Piccioli, M, Viezzoli, M.S. | | Deposit date: | 1998-04-24 | | Release date: | 1998-09-16 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced monomeric Q133M2 copper, zinc superoxide dismutase (SOD). Why is SOD a dimeric enzyme?.
Biochemistry, 37, 1998
|
|
1KVW
 
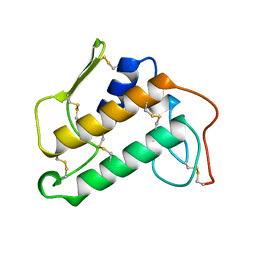 | |
3BBG
 
 | |
1BBG
 
 | |
2BBG
 
 | |
1BBX
 
 | | NON-SPECIFIC PROTEIN-DNA INTERACTIONS IN THE SSO7D-DNA COMPLEX, NMR, 1 STRUCTURE | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*CP*GP*CP*GP*CP*TP*AP*G)-3'), DNA-BINDING PROTEIN 7D | | Authors: | Agback, P, Baumann, H, Knapp, S, Ladenstein, R, Hard, T. | | Deposit date: | 1998-04-24 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Architecture of Nonspecific Protein-DNA Interactions in the Sso7D-DNA Complex
Nat.Struct.Biol., 5, 1998
|
|
1BBU
 
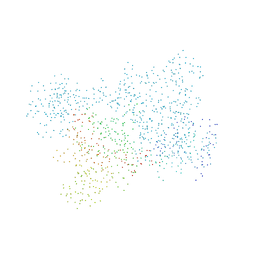 | | LYSYL-TRNA SYNTHETASE (LYSS) COMPLEXED WITH LYSINE | | Descriptor: | LYSINE, PROTEIN (LYSYL-TRNA SYNTHETASE) | | Authors: | Onesti, S, Desogus, G, Brevet, A, Chen, J, Plateau, P, Blanquet, S, Brick, P. | | Deposit date: | 1998-04-24 | | Release date: | 2000-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of lysyl-tRNA synthetase: conformational changes induced by substrate binding.
Biochemistry, 39, 2000
|
|
1NBE
 
 | |
1BBY
 
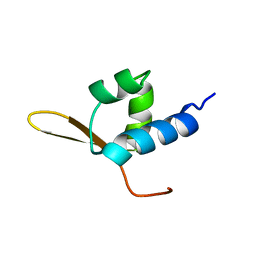 | | DNA-BINDING DOMAIN FROM HUMAN RAP30, NMR, MINIMIZED AVERAGE | | Descriptor: | RAP30 | | Authors: | Groft, C.M, Uljon, S.N, Wang, R, Werner, M.H. | | Deposit date: | 1998-04-26 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural homology between the Rap30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1C2N
 
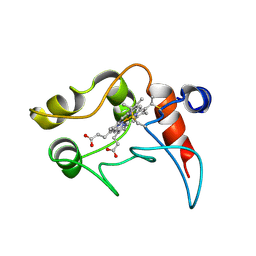 | | CYTOCHROME C2, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C2, HEME C | | Authors: | Cordier, F, Caffrey, M.S, Brutscher, B, Cusanovich, M.A, Marion, D, Blackledge, M. | | Deposit date: | 1998-04-27 | | Release date: | 1999-03-23 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure, rotational diffusion anisotropy and local backbone dynamics of Rhodobacter capsulatus cytochrome c2.
J.Mol.Biol., 281, 1998
|
|
2BBY
 
 | | DNA-BINDING DOMAIN FROM HUMAN RAP30, NMR, 30 STRUCTURES | | Descriptor: | RAP30 | | Authors: | Groft, C.M, Uljon, S.N, Wang, R, Werner, M.H. | | Deposit date: | 1998-04-27 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural homology between the Rap30 DNA-binding domain and linker histone H5: implications for preinitiation complex assembly.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1CA8
 
 | | Thrombin inhibitors with rigid tripeptidyl aldehydes | | Descriptor: | 2-{(3S)-3-[(benzylsulfonyl)amino]-2-oxopiperidin-1-yl}-N-{(2S)-1-[(3S)-1-carbamimidoylpiperidin-3-yl]-3-oxopropan-2-yl}acetamide, HIRUGEN, SODIUM ION, ... | | Authors: | Krishnan, R, Zhang, E, Hakansson, K, Arni, R.K, Tulinsky, A, Lim-Wilby, M.S.L, Levy, O.E, Semple, J.E, Brunck, T.K. | | Deposit date: | 1998-04-27 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes
Biochemistry, 37, 1998
|
|
1BB1
 
 | | CRYSTAL STRUCTURE OF A DESIGNED, THERMOSTABLE HETEROTRIMERIC COILED COIL | | Descriptor: | CHLORIDE ION, DESIGNED, THERMOSTABLE HETEROTRIMERIC COILED COIL | | Authors: | Nautiyal, S, Alber, T. | | Deposit date: | 1998-04-28 | | Release date: | 1999-02-02 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a designed, thermostable, heterotrimeric coiled coil.
Protein Sci., 8, 1999
|
|
1BB0
 
 | | THROMBIN INHIBITORS WITH RIGID TRIPEPTIDYL ALDEHYDES | | Descriptor: | 2-{(3S)-3-[(benzylsulfonyl)amino]-2-oxopiperidin-1-yl}-N-{(2S)-1-[(3R)-1-carbamimidoylpiperidin-3-yl]-3-oxopropan-2-yl}acetamide, HIRUGEN, SODIUM ION, ... | | Authors: | Krishnan, R, Zhang, E, Hakansson, K, Arni, R.K, Tulinsky, A, Lim-Wilby, M.S.L, Levy, O.E, Semple, J.E, Brunck, T.K. | | Deposit date: | 1998-04-28 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Highly selective mechanism-based thrombin inhibitors: structures of thrombin and trypsin inhibited with rigid peptidyl aldehydes.
Biochemistry, 37, 1998
|
|
1KVX
 
 | |
1RDR
 
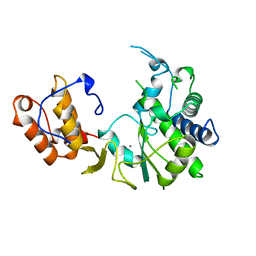 | | POLIOVIRUS 3D POLYMERASE | | Descriptor: | CALCIUM ION, POLIOVIRUS 3D POLYMERASE | | Authors: | Hansen, J, Long, A, Schultz, S. | | Deposit date: | 1998-04-28 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase of poliovirus.
Structure, 5, 1997
|
|
1BBZ
 
 | |
2PMT
 
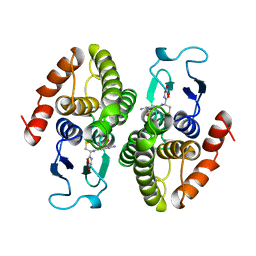 | | GLUTATHIONE TRANSFERASE FROM PROTEUS MIRABILIS | | Descriptor: | GLUTATHIONE, GLUTATHIONE TRANSFERASE | | Authors: | Rossjohn, J, Polekhina, G, Feil, S.C, Allocati, N, Masulli, M, Diilio, C, Parker, M.W. | | Deposit date: | 1998-04-28 | | Release date: | 1999-04-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A mixed disulfide bond in bacterial glutathione transferase: functional and evolutionary implications.
Structure, 6, 1998
|
|
1BB3
 
 | |
1BB4
 
 | |
1BB5
 
 | |
1BB9
 
 | |
1BCB
 
 | | INTRAMOLECULAR TRIPLEX, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*GP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*CP*T)-3') | | Authors: | Asensio, J.L, Brown, T, Lane, A.N. | | Deposit date: | 1998-04-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of intramolecular DNA triple helices containing adjacent and non-adjacent CG.C+ triplets.
Nucleic Acids Res., 26, 1998
|
|
