4Z7Y
 
 | | diphosphomevalonate decarboxylase from the Sulfolobus solfataricus, space group P21 | | Descriptor: | Diphosphomevalonate decarboxylase, SULFATE ION | | Authors: | Hattori, A, Unno, H, Hemmi, H. | | Deposit date: | 2015-04-08 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In Vivo Formation of the Protein Disulfide Bond That Enhances the Thermostability of Diphosphomevalonate Decarboxylase, an Intracellular Enzyme from the Hyperthermophilic Archaeon Sulfolobus solfataricus
J.Bacteriol., 197, 2015
|
|
4Z7C
 
 | | Diphosphomevalonate decarboxylase from the Sulfolobus solfataricus, space group h32 | | Descriptor: | Diphosphomevalonate decarboxylase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Unno, H, Hattori, A, Hemmi, H. | | Deposit date: | 2015-04-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In Vivo Formation of the Protein Disulfide Bond That Enhances the Thermostability of Diphosphomevalonate Decarboxylase, an Intracellular Enzyme from the Hyperthermophilic Archaeon Sulfolobus solfataricus
J.Bacteriol., 197, 2015
|
|
4UT4
 
 | | Burkholderia pseudomallei heptokinase WcbL, D-mannose complex. | | Descriptor: | CHLORIDE ION, PUTATIVE SUGAR KINASE, alpha-D-mannopyranose | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
4USK
 
 | | Unravelling the B. pseudomallei heptokinase WcbL: from Structure to Drug Discovery. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, PUTATIVE SUGAR KINASE, ... | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
4UTG
 
 | | Burkholderia pseudomallei heptokinase WcbL,AMPPNP (ATP analogue) complex. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-21 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
4USM
 
 | | WcbL complex with glycerol bound to sugar site | | Descriptor: | CHLORIDE ION, GLYCEROL, PUTATIVE SUGAR KINASE | | Authors: | Vivoli, M, Isupov, M.N, Nicholas, R, Hill, A, Scott, A, Kosma, P, Prior, J, Harmer, N.J. | | Deposit date: | 2014-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unraveling the B.Pseudomallei Heptokinase Wcbl: From Structure to Drug Discovery.
Chem.Biol., 22, 2015
|
|
5GMD
 
 | | Crystal structure of Sulfolobus solfataricus diphosphomevalonate decarboxylase in complex with ATP-gamma-S | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, ADENOSINE MONOPHOSPHATE, Diphosphomevalonate decarboxylase, ... | | Authors: | Unno, H, Hemmi, H, Hattori, A. | | Deposit date: | 2016-07-13 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Single Amino Acid Mutation Converts (R)-5-Diphosphomevalonate Decarboxylase into a Kinase
J. Biol. Chem., 292, 2017
|
|
5GME
 
 | | Crystal structure of Sulfolobus solfataricus Diphosphomevalonate decarboxylase in complex with ADP | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Unno, H, Hemmi, H, Hattori, A. | | Deposit date: | 2016-07-13 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Single Amino Acid Mutation Converts (R)-5-Diphosphomevalonate Decarboxylase into a Kinase
J. Biol. Chem., 292, 2017
|
|
5V2M
 
 | |
5V2L
 
 | |
5WAT
 
 | | Corynebacterium glutamicum Full length Homoserine kinase | | Descriptor: | HEXAETHYLENE GLYCOL, Homoserine kinase, MAGNESIUM ION, ... | | Authors: | Petit, C, Ronning, D.R. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Reduction of Feedback Inhibition in Homoserine Kinase (ThrB) ofCorynebacterium glutamicumEnhances l-Threonine Biosynthesis.
ACS Omega, 3, 2018
|
|
5WAS
 
 | | Corynebacterium glutamicum Hydrolyzed Homoserine kinase | | Descriptor: | Homoserine kinase, PHOSPHATE ION | | Authors: | Petit, C, Ronning, D.R. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Reduction of Feedback Inhibition in Homoserine Kinase (ThrB) ofCorynebacterium glutamicumEnhances l-Threonine Biosynthesis.
ACS Omega, 3, 2018
|
|
6CYZ
 
 | |
5YYS
 
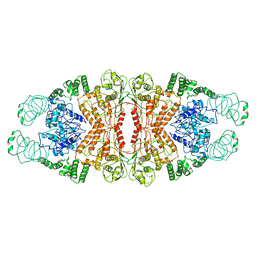 | | Cryo-EM structure of L-fucokinase, GDP-fucose pyrophosphorylase (FKP)in Bacteroides fragilis | | Descriptor: | L-fucokinase, L-fucose-1-P guanylyltransferase | | Authors: | Wang, J, Hu, H, Liu, Y, Zhou, Q, Wu, P, Yan, N, Wang, H.W, Wu, J.W, Sun, L. | | Deposit date: | 2017-12-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of L-fucokinase/GDP-fucose pyrophosphorylase (FKP) in Bacteroides fragilis.
Protein Cell, 10, 2019
|
|
6MDF
 
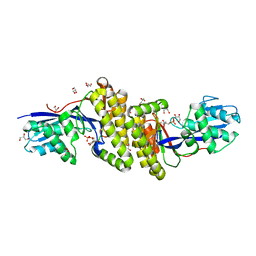 | |
6MDE
 
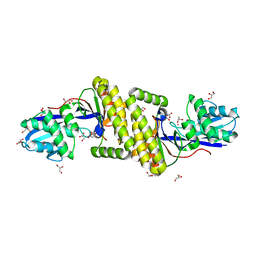 | |
6Q8Z
 
 | | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with N-(Cyclobutylmethyl)-1,5-dimethyl-1H-pyrazole-4-carboxamide
To Be Published
|
|
6Q91
 
 | | Structure of human galactokinase 1 bound with 5-Chloro-N-isobutyl-2-methoxybenzamide | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, 5-chloranyl-2-methoxy-~{N}-(2-methylpropyl)benzamide, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-17 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 5-Chloro-N-isobutyl-2-methoxybenzamide
To Be Published
|
|
6Q3W
 
 | | Structure of human galactokinase 1 bound with Ethyl 1-(2-pyrazinyl)-4-piperidinecarboxylate | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, beta-D-galactopyranose, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Structure of human galactokinase 1 bound with Ethyl 1-(2-pyrazinyl)-4-piperidinecarboxylate
To Be Published
|
|
6Q90
 
 | | Structure of human galactokinase 1 bound with 1-(4-Methoxyphenyl)-3-(4-pyridinyl)urea | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, Galactokinase, N-(4-methoxyphenyl)-N'-pyridin-4-ylurea, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2018-12-16 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 1-(4-Methoxyphenyl)-3-(4-pyridinyl)urea
To Be Published
|
|
6QJE
 
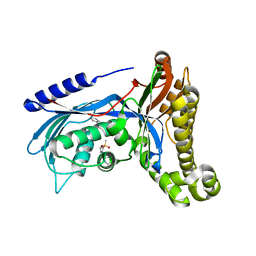 | | Structure of human galactokinase 1 bound with 4-{[2-(Methylsulfonyl)-1H-imidazol-1-yl]methyl}-1,3-thiazole | | Descriptor: | 2-(1,3-benzoxazol-2-ylamino)spiro[1,6,7,8-tetrahydroquinazoline-4,1'-cyclohexane]-5-one, 4-[(2-methylsulfonylimidazol-1-yl)methyl]-1,3-thiazole, Galactokinase, ... | | Authors: | Mackinnon, S.R, Bezerra, G.A, Zhang, M, Foster, W, Krojer, T, Brandao-Neto, J, Douangamath, A, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, Lai, K, Yue, W.W. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human galactokinase 1 bound with 4-{[2-(Methylsulfonyl)-1H-imidazol-1-yl]methyl}-1,3-thiazole
To Be Published
|
|
6GR2
 
 | | Structure of human galactokinase in complex with galactose and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Galactokinase, SULFATE ION, ... | | Authors: | Bezerra, G.A, Mackinnon, S, Williams, E, Zhang, M, Arrowsmith, C, Edwards, A, Bountra, C, Lai, K, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-08 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of human galactokinase in complex with galactose and ADP
To Be Published
|
|
6N10
 
 | | Crystal structure of Arabidopsis thaliana mevalonate 5-diphosphate decarboxylase 1 complexed with (R)-MVAPP | | Descriptor: | (3R)-3-HYDROXY-5-{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}-3-METHYLPENTANOIC ACID, Diphosphomevalonate decarboxylase MVD1, peroxisomal, ... | | Authors: | Noel, J.P, Thomas, S.T, Louie, G.V. | | Deposit date: | 2018-11-07 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Specificity and Engineering of Mevalonate 5-Phosphate Decarboxylase.
Acs Chem.Biol., 14, 2019
|
|
6JBC
 
 | | Phosphotransferase related to CoA biosynthesis pathway | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Kita, A, Kishimoto, A, Shimosaka, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pantoate kinase from Thermococcus kodakarensis.
Proteins, 88, 2020
|
|
6JBD
 
 | | Phosphotransferase-ATP complex related to CoA biosynthesis pathway | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Kita, A, Kishimoto, A, Shimosaka, T, Tomita, H, Yokooji, Y, Imanaka, T, Atomi, H, Miki, K. | | Deposit date: | 2019-01-25 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of pantoate kinase from Thermococcus kodakarensis.
Proteins, 88, 2020
|
|
