1M7Q
 
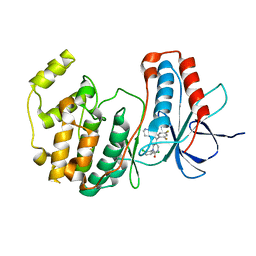 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | Deposit date: | 2002-07-22 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
2P3I
 
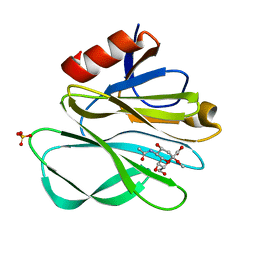 | | Crystal structure of Rhesus Rotavirus VP8* at 295K | | Descriptor: | 2-O-methyl-5-N-acetyl-alpha-D-neuraminic acid, SULFATE ION, VP4 | | Authors: | Blanchard, H. | | Deposit date: | 2007-03-09 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Effects on sialic acid recognition of amino acid mutations in the carbohydrate-binding cleft of the rotavirus spike protein
Glycobiology, 19, 2009
|
|
1M5V
 
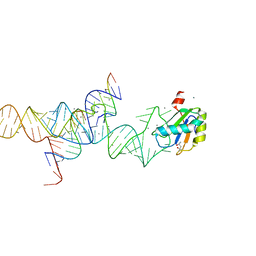 | | Transition State Stabilization by a Catalytic RNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, RNA HAIRPIN RIBOZYME, ... | | Authors: | Rupert, P.B, Massey, A.P, Sigurdsson, S.T, Ferre-D'Amare, A.R. | | Deposit date: | 2002-07-09 | | Release date: | 2002-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Transition state stabilization by a catalytic RNA
Science, 298, 2002
|
|
7RVP
 
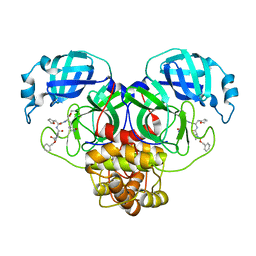 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI14 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-furan-2-yl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
9GBJ
 
 | | KRAS G12D in complex with covalent inhibitor | | Descriptor: | 1-[(3S)-1-[2-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]-6-[(1S)-1-[(2S)-1-methylpyrrolidin-2-yl]ethoxy]pyrimidin-4-yl]pyrrolidin-3-yl]-3-[1-(methoxymethyl)cyclopropyl]urea, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Sirocchi, L.S, Zak, K.M, Wilding, B. | | Deposit date: | 2024-07-31 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Discovery of Carbodiimide Warheads to Selectively and Covalently Target Aspartic Acid in KRAS G12D .
J.Am.Chem.Soc., 147, 2025
|
|
7RVN
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI12 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methylidene-L-norvalinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
1M69
 
 | |
7RVY
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI25 | | Descriptor: | 3C-like proteinase, O-tert-butyl-N-{[(3-chlorophenyl)methoxy]carbonyl}-L-threonyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RJ9
 
 | |
1M8P
 
 | |
9FYU
 
 | |
7RHA
 
 | | A new fluorescent protein darkmRuby at pH 5.0 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Huang, M, Ng, H.L, Zhang, S, Deng, M, Chu, J. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a new fluorescent protein darkmRuby at pH 5.0
To Be Published
|
|
9GS1
 
 | |
1M6U
 
 | | Crystal Structure of a Novel DNA-binding domain from Ndt80, a Transcriptional Activator Required for Meiosis in Yeast | | Descriptor: | Ndt80 protein, SULFATE ION | | Authors: | Montano, S.P, Cote, M.L, Fingerman, I, Pierce, M, Vershon, A.K, Georgiadis, M.M. | | Deposit date: | 2002-07-17 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the DNA-binding domain from Ndt80, a transcriptional activator required for meiosis in yeast
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
7RVT
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI20 | | Descriptor: | 3C-like proteinase, N~2~-[(2S)-2-{[(benzyloxy)carbonyl]amino}-2-cyclopropylacetyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-4-methyl-L-leucinamide | | Authors: | Yang, K, Sankaran, B, Liu, W. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals.
Eur.J.Med.Chem., 240, 2022
|
|
7RE9
 
 | | TCR mimic antibody (Fab fragment) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fab heavy chain, Fab light chain | | Authors: | Dasgupta, M, Baker, B.M. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Validation and promise of a TCR mimic antibody for cancer immunotherapy of hepatocellular carcinoma.
Sci Rep, 12, 2022
|
|
9GS5
 
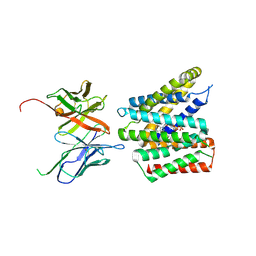 | | Cryo-EM structure of human SLC35B1-E33A variant with ADP in outward facing conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Fv-MBP, Solute carrier family 35 member B1 | | Authors: | Gulati, A, Ahn, D, Suades, A, Drew, D. | | Deposit date: | 2024-09-13 | | Release date: | 2025-05-21 | | Last modified: | 2025-08-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise ATP translocation into the endoplasmic reticulum by human SLC35B1.
Nature, 643, 2025
|
|
1M7H
 
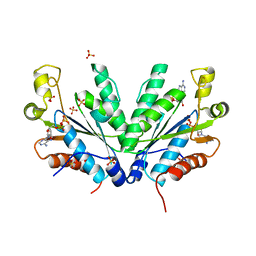 | | Crystal Structure of APS kinase from Penicillium Chrysogenum: Structure with APS soaked out of one dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-PHOSPHOSULFATE, Adenylylsulfate kinase, ... | | Authors: | Lansdon, E.B, Sege, I.H, Fisher, A.J. | | Deposit date: | 2002-07-19 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand-Induced Structural Changes in Adenosine 5'-Phosphosulfate
Kinase from Penicillium chrysogenum.
Biochemistry, 41, 2002
|
|
9GKN
 
 | |
9GSZ
 
 | |
1M9Z
 
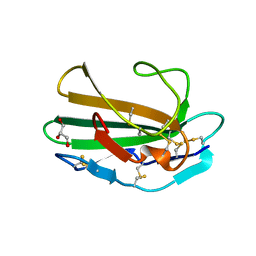 | | CRYSTAL STRUCTURE OF HUMAN TGF-BETA TYPE II RECEPTOR LIGAND BINDING DOMAIN | | Descriptor: | GLYCEROL, TGF-BETA RECEPTOR TYPE II | | Authors: | Boesen, C.C, Radaev, S, Motyka, S.A, Patamawenu, A, Sun, P.D. | | Deposit date: | 2002-07-30 | | Release date: | 2002-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | THE 1.1A CRYSTAL STRUCTURE OF HUMAN TGF-BETA TYPE II RECEPTOR LIGAND BINDING DOMAIN
Structure, 10, 2002
|
|
2PD3
 
 | | Crystal Structure of the Helicobacter pylori Enoyl-Acyl Carrier Protein Reductase in Complex with Hydroxydiphenyl Ether Compounds, Triclosan and Diclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Lee, H.H, Moon, J.H, Suh, S.W. | | Deposit date: | 2007-03-31 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the Helicobacter pylori enoyl-acyl carrier protein reductase in complex with hydroxydiphenyl ether compounds, triclosan and diclosan
Proteins, 69, 2007
|
|
1MA1
 
 | | Structure and properties of the atypical iron superoxide dismutase from Methanobacterium thermoautotrophicum | | Descriptor: | FE (III) ION, superoxide dismutase | | Authors: | Adams, J.J, Anderson, B.F, Renault, J.P, Verchere-Beaur, C, Morgenstern-Badarau, I, Jameson, G.B. | | Deposit date: | 2002-07-31 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and properties of the atypical iron superoxide dismutase from Methanobacterium thermoautotrophicum
To be published
|
|
7REB
 
 | | E. coli dihydrofolate reductase complexed with 5-(3-(7-(4-(aminomethyl)phenyl)benzo[d][1,3]dioxol-5-yl)but-1-yn-1-yl)-6-ethylpyrimidine-2,4-diamine (UCP1223) | | Descriptor: | 5-[(3R)-3-{7-[4-(aminomethyl)phenyl]-2H-1,3-benzodioxol-5-yl}but-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Lombardo, M.N, Wright, D.L. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens.
Commun Biol, 5, 2022
|
|
7RHB
 
 | |
