1AD1
 
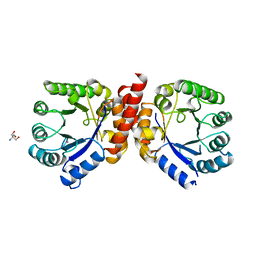 | | DIHYDROPTEROATE SYNTHETASE (APO FORM) FROM STAPHYLOCOCCUS AUREUS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIHYDROPTEROATE SYNTHETASE, POTASSIUM ION | | Authors: | Kostrewa, D, Oefner, C, D'Arcy, A. | | Deposit date: | 1997-02-19 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the dihydropteroate synthase from Staphylococcus aureus.
J.Mol.Biol., 268, 1997
|
|
7GAB
 
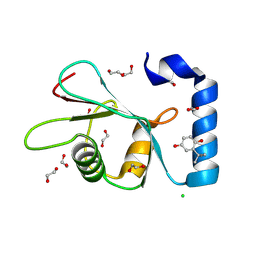 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z1255402624 | | Descriptor: | 1,2-ETHANEDIOL, 2-tert-butylbenzene-1,4-diol, CHLORIDE ION, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4K0Z
 
 | |
8E5B
 
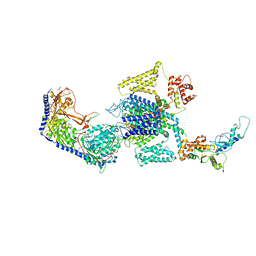 | | Human L-type voltage-gated calcium channel Cav1.3 in the presence of Amiodarone and Sofosbuvir at 3.3 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
7GAM
 
 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z373768900 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Microtubule-associated proteins 1A/1B light chain 3B, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4CUS
 
 | | Crystal structure of human BAZ2B in complex with fragment-4 N09496 | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2B, quinolin-4-ol | | Authors: | Bradley, A.R, Liu, Y, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, von Delft, F. | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Crystal Structure of Human Baz2B in Complex with Fragment-4 N09496
To be Published
|
|
3URZ
 
 | |
7GAQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z755044716 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Microtubule-associated proteins 1A/1B light chain 3B, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
8E5A
 
 | | Human L-type voltage-gated calcium channel Cav1.3 treated with 1.4 mM Sofosbuvir at 3.3 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
2O99
 
 | | The crystal structure of E.coli IclR C-terminal fragment in complex with glyoxylate | | Descriptor: | 1,2-ETHANEDIOL, Acetate operon repressor, GLYCOLIC ACID | | Authors: | Lunin, V.V, Ezersky, A, Evdokimova, E, Kudritska, M, Savchenko, A. | | Deposit date: | 2006-12-13 | | Release date: | 2007-04-10 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Glyoxylate and Pyruvate Are Antagonistic Effectors of the Escherichia coli IclR Transcriptional Regulator.
J.Biol.Chem., 282, 2007
|
|
6QTP
 
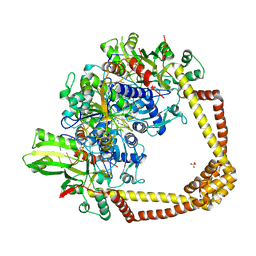 | | 2.37A structure of gepotidacin with S.aureus DNA gyrase and uncleaved DNA | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*AP*GP*CP*TP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Bax, B.D. | | Deposit date: | 2019-02-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanistic and Structural Basis for the Actions of the Antibacterial Gepotidacin against Staphylococcus aureus Gyrase.
Acs Infect Dis., 5, 2019
|
|
7GAE
 
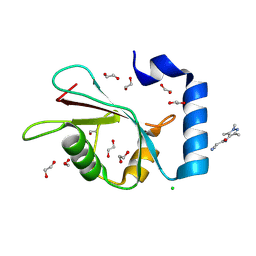 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z1688504114 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyano-~{N}-(1,3,5-trimethylpyrazol-4-yl)ethanamide, CHLORIDE ION, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1YYC
 
 | |
6ATF
 
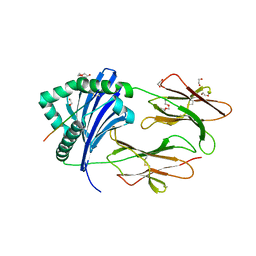 | | HLA-DRB1*1402 in complex with Vimentin59-71 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scally, S.W, Ting, Y.T, Rossjohn, J. | | Deposit date: | 2017-08-29 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for increased susceptibility of Indigenous North Americans to seropositive rheumatoid arthritis.
Ann. Rheum. Dis., 76, 2017
|
|
3I7A
 
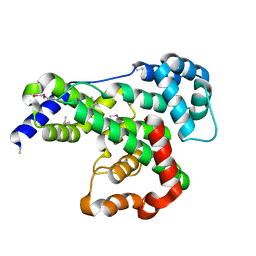 | |
5HZA
 
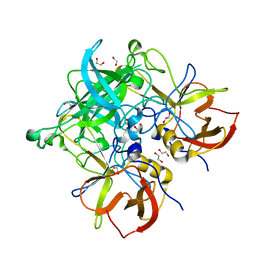 | | Crystal structure of GII.10 P domain in complex with 3-fucosyllactose (3 FL) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein, alpha-L-fucopyranose-(1-3)-[beta-D-galactopyranose-(1-4)]beta-D-glucopyranose | | Authors: | Hansman, G.S, Koromyslova, A.D, Singh, B.K. | | Deposit date: | 2016-02-02 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Basis for Norovirus Inhibition by Human Milk Oligosaccharides.
J.Virol., 90, 2016
|
|
1AHN
 
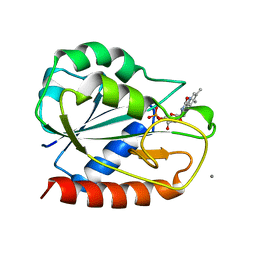 | | E. COLI FLAVODOXIN AT 2.6 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Ludwig, M.L. | | Deposit date: | 1997-04-07 | | Release date: | 1997-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A flavodoxin that is required for enzyme activation: the structure of oxidized flavodoxin from Escherichia coli at 1.8 A resolution.
Protein Sci., 6, 1997
|
|
6B1F
 
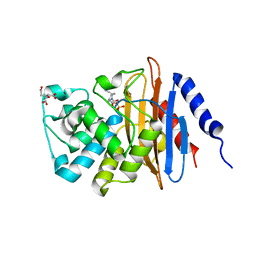 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 4234 by soaking | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carbonitrile, 1,2-ETHANEDIOL, CITRIC ACID, ... | | Authors: | van den Akker, F, Nguyen, N.Q. | | Deposit date: | 2017-09-18 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
5TNS
 
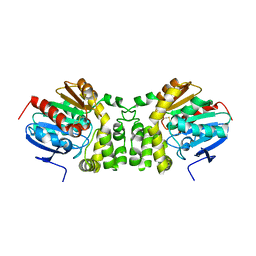 | |
6HW3
 
 | | Yeast 20S proteasome in complex with 13 | | Descriptor: | (2~{S})-~{N}-[(2~{S})-1-[[(2~{S})-1-[4-(aminomethyl)phenyl]-4-methylsulfonyl-butan-2-yl]amino]-3-oxidanyl-1-oxidanylidene-propan-2-yl]-2-[[(2~{S})-2-azido-3-phenyl-propanoyl]amino]-4-methyl-pentanamide, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2018-10-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Inhibitors Selective for Human Proteasome beta 2c or beta 2i Subunits.
J.Med.Chem., 62, 2019
|
|
1A8Z
 
 | | STRUCTURE DETERMINATION OF A 16.8KDA COPPER PROTEIN RUSTICYANIN AT 2.1A RESOLUTION USING ANOMALOUS SCATTERING DATA WITH DIRECT METHODS | | Descriptor: | COPPER (I) ION, RUSTICYANIN | | Authors: | Harvey, I, Hao, Q, Duke, E.M.H, Ingledew, W.J, Hasnain, S.S. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure determination of a 16.8 kDa copper protein at 2.1 A resolution using anomalous scattering data with direct methods.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
5Y02
 
 | | C-terminal peptide depleted mutant of hydroxynitrile lyase from Passiflora edulis (PeHNL) bound with (R)-mandelonitrile | | Descriptor: | (2R)-hydroxy(phenyl)ethanenitrile, HEXANE-1,6-DIOL, Hydroxynitrile lyase, ... | | Authors: | Motojima, F, Nuylert, A, Asano, Y. | | Deposit date: | 2017-07-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure and catalytic mechanism of hydroxynitrile lyase from passion fruit, Passiflora edulis
FEBS J., 285, 2018
|
|
5CS3
 
 | | The structure of the NK1 fragment of HGF/SF complexed with (H)EPPS | | Descriptor: | 3-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]PROPANE-1-SULFONIC ACID, Hepatocyte growth factor | | Authors: | Sigurdardottir, A.G, Winter, A, Sobkowicz, A, Fragai, M, Chirgadze, D.Y, Ascher, D.B, Blundell, T.L, Gherardi, E. | | Deposit date: | 2015-07-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Exploring the chemical space of the lysine-binding pocket of the first kringle domain of hepatocyte growth factor/scatter factor (HGF/SF) yields a new class of inhibitors of HGF/SF-MET binding.
Chem Sci, 6, 2015
|
|
3IDO
 
 | |
4CUT
 
 | | Crystal structure of human BAZ2B in complex with fragment-5 N09428 | | Descriptor: | 1,2-ETHANEDIOL, BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2B, N-(4-HYDROXYPHENYL)ACETAMIDE (TYLENOL) | | Authors: | Bradley, A.R, Liu, Y, Krojer, T, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, von Delft, F. | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Crystal Structure of Human Baz2B in Complex with Fragment-5 N09428
To be Published
|
|
