7DGW
 
 | | De novo designed protein H4A2S | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, de novo designed protein H4A2S | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DGU
 
 | | De novo designed protein H4A1R | | Descriptor: | de novo designed protein H4A1R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DKK
 
 | | De novo design protein XM2H | | Descriptor: | De novo design protein XM2H | | Authors: | Bin, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DKO
 
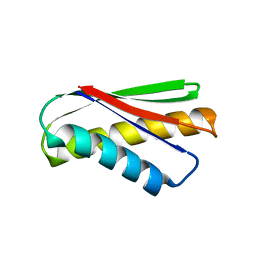 | | De novo design protein AM2M | | Descriptor: | de novo designed protein AM2M | | Authors: | Bin, H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7DMF
 
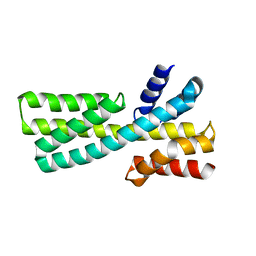 | |
7DGY
 
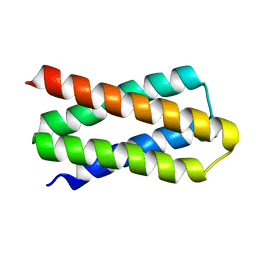 | | De novo designed protein H4C2R | | Descriptor: | de novo designed protein H4C2R | | Authors: | Xu, Y, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2020-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A backbone-centred energy function of neural networks for protein design.
Nature, 602, 2022
|
|
7NEI
 
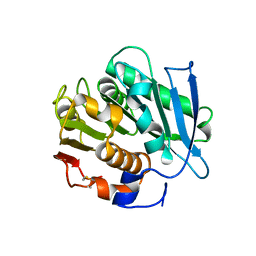 | |
6YKR
 
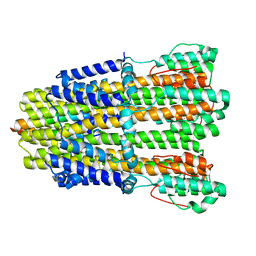 | |
6YKP
 
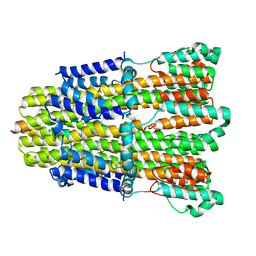 | |
6GCT
 
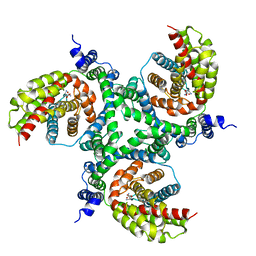 | | cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Garaeva, A.A, Oostergetel, G.T, Gati, C, Guskov, A, Paulino, C, Slotboom, D.J. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM structure of the human neutral amino acid transporter ASCT2.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6Y3D
 
 | | X-ray structure of thermophilic C-phycocyanin from Galdiera phlegrea | | Descriptor: | ACETATE ION, C-phycocyanin alpha chain, C-phycocyanin beta chain, ... | | Authors: | Ferraro, G, Lucignano, R, Marseglia, A, Merlino, A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of C-phycocyanin from Galdieria phlegrea: Determinants of thermostability and comparison with a C-phycocyanin in the entire phycobilisome.
Biochim Biophys Acta Bioenerg, 1861, 2020
|
|
6YKM
 
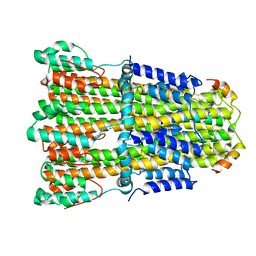 | | Structure of C. jejuni MotAB | | Descriptor: | Chemotaxis protein MotA, putative, Chemotaxis protein MotB | | Authors: | Santiveri, M, Roa-Eguiara, A, Taylor, N.M.I. | | Deposit date: | 2020-04-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and Function of Stator Units of the Bacterial Flagellar Motor.
Cell, 183, 2020
|
|
6UT1
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH BNM-III-170 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HIV-1 LM/HS clade A/E CRF01 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
8EHS
 
 | |
8EHR
 
 | |
8EHT
 
 | |
1GKN
 
 | | Structure Determination and Rational Mutagenesis reveal binding surface of immune adherence receptor, CR1 (CD35) | | Descriptor: | COMPLEMENT RECEPTOR TYPE 1 | | Authors: | Smith, B.O, Mallin, R.L, Krych-Goldberg, M, Wang, X, Hauhart, R.E, Bromek, K, Uhrin, D, Atkinson, J.P, Barlow, P.N. | | Deposit date: | 2001-08-16 | | Release date: | 2002-04-18 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C3B Binding Site of Cr1 (Cd35), the Immune Adherence Receptor
Cell(Cambridge,Mass.), 108, 2002
|
|
7SXM
 
 | |
1PCQ
 
 | | Crystal structure of groEL-groES | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Chaudhry, C, Farr, G.W, Todd, M.J, Rye, H.S, Brunger, A.T, Adams, P.D, Horwich, A.L, Sigler, P.B. | | Deposit date: | 2003-05-16 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics.
Embo J., 22, 2003
|
|
1PF9
 
 | | GroEL-GroES-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, groEL protein, ... | | Authors: | Chaudhry, C, Farr, G.W, Todd, M.J, Rye, H.S, Brunger, A.T, Adams, P.D, Horwich, A.L, Sigler, P.B. | | Deposit date: | 2003-05-24 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Role of the gamma-phosphate of ATP in triggering protein folding by GroEL-GroES: function, structure and energetics.
Embo J., 22, 2003
|
|
3NQ8
 
 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution R4 8/5A | | Descriptor: | BENZAMIDINE, NITRATE ION, deoxyribose phosphate aldolase | | Authors: | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2010-06-29 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
6REZ
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form, pH 5.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6WIY
 
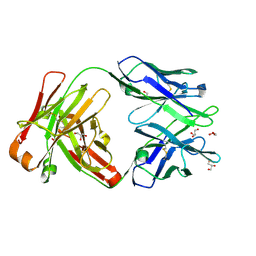 | | Crystal structure of Fab 54-1G05 | | Descriptor: | Fab 54-1G05 heavy chain, Fab 54-1G05 light chain, GLYCEROL | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
6REX
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric form, pH 6.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6WIZ
 
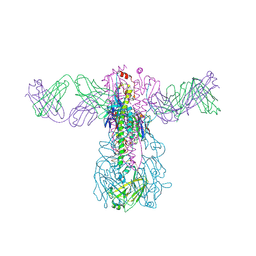 | | Crystal structure of Fab 54-1G05 bound to H1 influenza hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 54-1G05 heavy chain, Fab 54-1G05 light chain, ... | | Authors: | Wu, N.C, Wilson, I.A. | | Deposit date: | 2020-04-11 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Convergent Evolution in Breadth of Two VH6-1-Encoded Influenza Antibody Clonotypes from a Single Donor.
Cell Host Microbe, 28, 2020
|
|
