1KW7
 
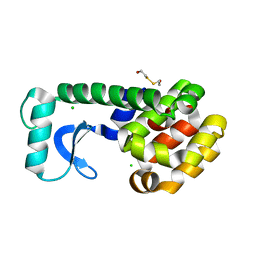 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-01-28 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
1KWS
 
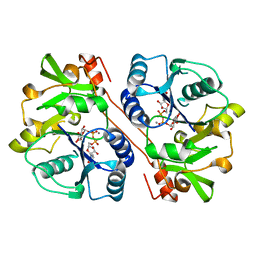 | | CRYSTAL STRUCTURE OF BETA1,3-GLUCURONYLTRANSFERASE I IN COMPLEX WITH THE ACTIVE UDP-GLCUA DONOR | | Descriptor: | BETA-1,3-GLUCURONYLTRANSFERASE 3, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Pedersen, L.C, Darden, T.A, Negishi, M. | | Deposit date: | 2002-01-30 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta 1,3-glucuronyltransferase I in complex with active donor substrate UDP-GlcUA.
J.Biol.Chem., 277, 2002
|
|
1KRW
 
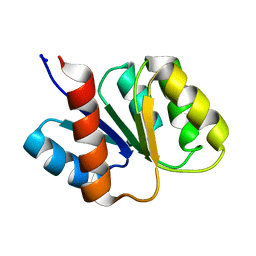 | | SOLUTION STRUCTURE AND BACKBONE DYNAMICS OF BERYLLOFLUORIDE-ACTIVATED NTRC RECEIVER DOMAIN | | Descriptor: | NITROGEN REGULATION PROTEIN NR(I) | | Authors: | Hastings, C.A, Lee, S.-Y, Cho, H.S, Yan, D, Kustu, S, Wemmer, D.E. | | Deposit date: | 2002-01-10 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Solution Structure of the Beryllofluoride-Activated NtrC Receiver Domain
Biochemistry, 42, 2003
|
|
7A9F
 
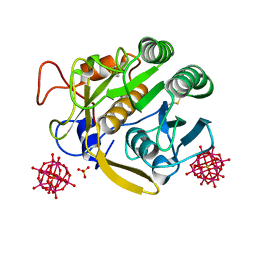 | |
1KU1
 
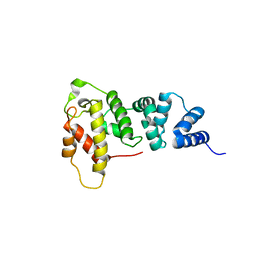 | | Crystal Structure of the Sec7 Domain of Yeast GEA2 | | Descriptor: | ARF guanine-nucleotide exchange factor 2 | | Authors: | Renault, L, Christova, P, Guibert, B, Pasqualato, S, Cherfils, J. | | Deposit date: | 2002-01-20 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism of domain closure of Sec7 domains and role in BFA sensitivity.
Biochemistry, 41, 2002
|
|
1KVT
 
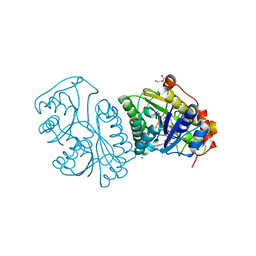 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular structures of the S124A, S124T, and S124V site-directed mutants of UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
7A9M
 
 | |
1KOD
 
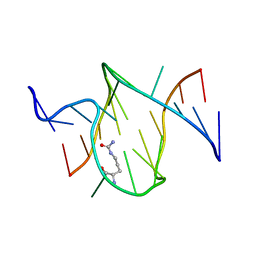 | | RNA APTAMER COMPLEXED WITH CITRULLINE, NMR | | Descriptor: | CITRULLINE, RNA (5'-R(P*AP*CP*GP*GP*UP*UP*AP*GP*GP*UP*CP*GP*CP*U)-3'), RNA (5'-R(P*AP*GP*AP*AP*GP*GP*AP*GP*UP*GP*U)-3') | | Authors: | Yang, Y.S, Kochoyan, M, Burgstaller, P, Westhof, E, Famulok, M. | | Deposit date: | 1996-03-28 | | Release date: | 1996-11-08 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy.
Science, 272, 1996
|
|
9CBT
 
 | | Crystal structure of human sirtuin 3 fragment (residues 118-399) bound to intermediates from reaction with NAD and inhibitor NH6-10 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-6-[[(2~{R},3~{a}~{R},5~{R},6~{R},6~{a}~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-6-oxidanyl-2-tridecyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]amino]-1-oxidanylidene-1-[2-(triethyl-$l^{4}-azanyl)ethylamino]hexan-2-yl]carbamate, 2-{[(2S)-6-[(Z)-(1-{[(2R,3R,4R,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-4-hydroxy-2-sulfanyloxolan-3-yl]oxy}tetradecylidene)amino]-2-{[(benzyloxy)carbonyl]amino}hexanoyl]amino}-N,N,N-triethylethan-1-aminium (non-preferred name), NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Fenwick, M.K, Young, H.J, Lin, H. | | Deposit date: | 2024-06-20 | | Release date: | 2024-08-07 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Mitochondria-Targeting SIRT3 Inhibitor with Activity against Diffuse Large B Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
1KDD
 
 | | X-ray structure of the coiled coil GCN4 ACID BASE HETERODIMER ACID-d12La16I BASE-d12La16L | | Descriptor: | GCN4 ACID BASE HETERODIMER ACID-d12La16I, GCN4 ACID BASE HETERODIMER BASE-d12La16L | | Authors: | Keating, A.E, Malashkevich, V.N, Tidor, B, Kim, P.S. | | Deposit date: | 2001-11-12 | | Release date: | 2001-11-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Side-chain repacking calculations for predicting structures and stabilities of heterodimeric coiled coils.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7A18
 
 | | 50S Deinococcus radiodurans ribosome bounded with mycinamicin IV | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L15, ... | | Authors: | Breiner, E, Eyal, Z, Matzov, D, Halfon, Y, Cimicata, G, Rozenberg, H, Zimmerman, E, Bashan, A, Yonath, A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ribosome-binding and anti-microbial studies of the mycinamicins, 16-membered macrolide antibiotics from Micromonospora griseorubida.
Nucleic Acids Res., 49, 2021
|
|
1KDK
 
 | | THE STRUCTURE OF THE N-TERMINAL LG DOMAIN OF SHBG IN CRYSTALS SOAKED WITH EDTA | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Sex Hormone-Binding Globulin | | Authors: | Grishkovskaya, I, Avvakumov, G.V, Hammond, G.L, Muller, Y.A. | | Deposit date: | 2001-11-13 | | Release date: | 2002-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Resolution of a disordered region at the entrance of the human sex hormone-binding globulin steroid-binding site.
J.Mol.Biol., 318, 2002
|
|
1KV8
 
 | | Crystal Structure of 3-Keto-L-Gulonate 6-Phosphate Decarboxylase | | Descriptor: | 3-Keto-L-Gulonate 6-Phosphate Decarboxylase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Wise, E, Yew, W.S, Babbitt, P.C, Gerlt, J.A, Rayment, I. | | Deposit date: | 2002-01-25 | | Release date: | 2002-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Homologous (beta/alpha)8-barrel enzymes that catalyze unrelated reactions: orotidine 5'-monophosphate decarboxylase and 3-keto-L-gulonate 6-phosphate decarboxylase.
Biochemistry, 41, 2002
|
|
7AD5
 
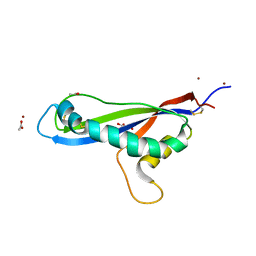 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | Descriptor: | ACETATE ION, Avirulence protein LmJ1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
1KDR
 
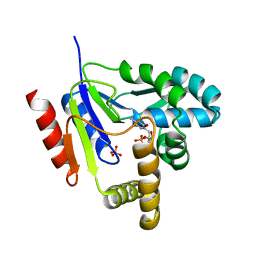 | | CYTIDINE MONOPHOSPHATE KINASE FROM E.COLI IN COMPLEX WITH ARA-CYTIDINE MONOPHOSPHATE | | Descriptor: | CYTIDYLATE KINASE, CYTOSINE ARABINOSE-5'-PHOSPHATE, SULFATE ION | | Authors: | Bertrand, T, Briozzo, P, Assairi, L, Ofiteru, A, Bucurenci, N, Munier-Lehmann, H, Golinelli-Pimpaneau, B, Barzu, O, Gilles, A.M. | | Deposit date: | 2001-11-13 | | Release date: | 2002-01-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme.
J.Mol.Biol., 315, 2002
|
|
1KE6
 
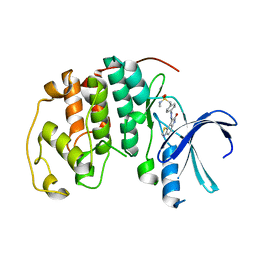 | | CYCLIN-DEPENDENT KINASE 2 (CDK2) COMPLEXED WITH N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Descriptor: | Cell division protein kinase 2, N-METHYL-{4-[2-(7-OXO-6,7-DIHYDRO-8H-[1,3]THIAZOLO[5,4-E]INDOL-8-YLIDENE)HYDRAZINO]PHENYL}METHANESULFONAMIDE | | Authors: | Bramson, H.N, Corona, J, Davis, S.T, Dickerson, S.H, Edelstein, M, Frye, S.V, Gampe, R.T, Hassell, A.H, Shewchuk, L.M, Kuyper, L.F. | | Deposit date: | 2001-11-14 | | Release date: | 2002-05-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oxindole-based inhibitors of cyclin-dependent kinase 2 (CDK2): design, synthesis, enzymatic activities, and X-ray crystallographic analysis.
J.Med.Chem., 44, 2001
|
|
1KW6
 
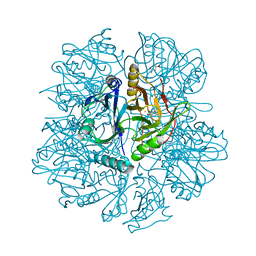 | | Crystal structure of 2,3-dihydroxybiphenyl dioxygenase (BphC) in complex with 2,3-dihydroxybiphenyl at 1.45 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2,3-Dihydroxybiphenyl dioxygenase, BIPHENYL-2,3-DIOL, ... | | Authors: | Sato, N, Uragami, Y, Nishizaki, T, Takahashi, Y, Sazaki, G, Sugimoto, K, Nonaka, T, Masai, E, Fukuda, M, Senda, T. | | Deposit date: | 2002-01-28 | | Release date: | 2003-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of the Reaction Intermediate and its Homologue of an Extradiol-cleaving Catecholic Dioxygenase
J.Mol.Biol., 321, 2002
|
|
6P7S
 
 | | Crystal Structure of the Cedar henipavirus Attachment G Glycoprotein globular domain in complex with the receptor ephrin-B1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Attachment glycoprotein, ... | | Authors: | Xu, K, Nikolov, D.B, Xu, Y. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structural and functional analyses reveal promiscuous and species specific use of ephrin receptors by Cedar virus.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1KEC
 
 | | PENICILLIN ACYLASE MUTANT WITH PHENYL PROPRIONIC ACID | | Descriptor: | CALCIUM ION, PENICILLIN ACYLASE ALPHA SUBUNIT, PENICILLIN ACYLASE BETA SUBUNIT, ... | | Authors: | Hensgens, C.M.H, Keizer, E, Snijder, H.J, Dijkstra, B.W. | | Deposit date: | 2001-11-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies on ligand binding in wild-type and active-site mutants of penicillin acylase.
Protein Eng.Des.Sel., 17, 2004
|
|
1KET
 
 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with thymidine diphosphate bound | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, THYMIDINE-5'-DIPHOSPHATE, dTDP-D-glucose 4,6-dehydratase | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
1KF2
 
 | | Atomic Resolution Structure of RNase A at pH 5.2 | | Descriptor: | SULFATE ION, pancreatic ribonuclease | | Authors: | Berisio, R, Sica, F, Lamzin, V.S, Wilson, K.S, Zagari, A, Mazzarella, L. | | Deposit date: | 2001-11-19 | | Release date: | 2001-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic resolution structures of ribonuclease A at six pH values.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KX2
 
 | | Minimized average structure of a mono-heme ferrocytochrome c from Shewanella putrefaciens | | Descriptor: | HEME C, mono-heme c-type cytochrome ScyA | | Authors: | Bartalesi, I, Bertini, I, Hajieva, P, Rosato, A, Vasos, P.R. | | Deposit date: | 2002-01-30 | | Release date: | 2002-02-13 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a monoheme ferrocytochrome c from Shewanella putrefaciens and structural analysis of sequence-similar proteins: functional implications.
Biochemistry, 41, 2002
|
|
1KX9
 
 | | ANTENNAL CHEMOSENSORY PROTEIN A6 FROM THE MOTH MAMESTRA BRASSICAE | | Descriptor: | ACETATE ION, CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and ligand binding study of a moth chemosensory protein
J.Biol.Chem., 277, 2002
|
|
1KF9
 
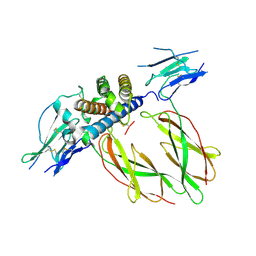 | | PHAGE DISPLAY DERIVED VARIANT OF HUMAN GROWTH HORMONE COMPLEXED WITH TWO COPIES OF THE EXTRACELLULAR DOMAIN OF ITS RECEPTOR | | Descriptor: | EXTRACELLULAR DOMAIN HUMAN GROWTH HORMONE RECEPTOR (1-238), PHAGE DISPLAY DERIVED VARIANT HUMAN GROWTH HORMONE | | Authors: | Schiffer, C.A, Ultsch, M, Walsh, S, Somers, W, De Vos, A.M, Kossiakoff, A.A. | | Deposit date: | 2001-11-19 | | Release date: | 2002-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a Phage Display Derived Variant of Human Growth Hormone Complexed to Two Copies of the Extracellular Domain of its Receptor: Evidence for Strong Structural Coupling between Receptor Binding Sites
J.Mol.Biol., 316, 2002
|
|
1KFT
 
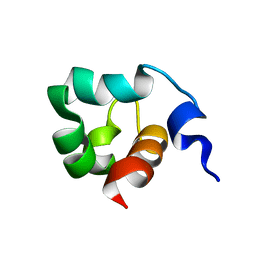 | | Solution Structure of the C-Terminal domain of UvrC from E-coli | | Descriptor: | Excinuclease ABC subunit C | | Authors: | Singh, S, Folkers, G.E, Bonvin, A.M.J.J, Boelens, R, Wechselberger, R, Niztayev, A, Kaptein, R. | | Deposit date: | 2001-11-23 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E.coli
EMBO J., 21, 2002
|
|
