6RNX
 
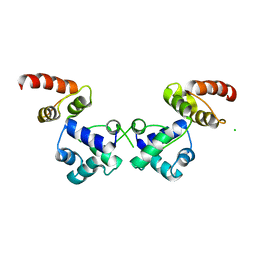 | | Crystal structure of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | CHLORIDE ION, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
6RMQ
 
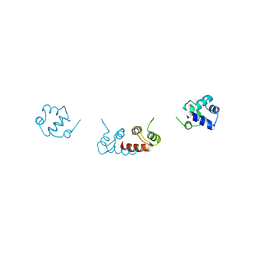 | | Crystal structure of a selenomethionine-substituted A70M I84M mutant of the essential repressor DdrO from radiation resistant-Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-07 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
6LTY
 
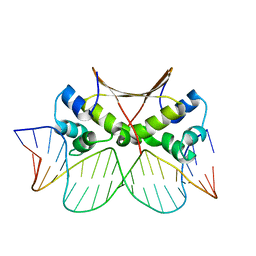 | | DNA bound antitoxin HigA3 | | Descriptor: | DNA (5'-D(P*CP*CP*AP*CP*GP*AP*GP*AP*TP*AP*TP*AP*AP*CP*CP*TP*AP*GP*AP*G)-3'), DNA (5'-D(P*CP*TP*CP*TP*AP*GP*GP*TP*TP*AP*TP*AP*TP*CP*TP*CP*GP*TP*GP*G)-3'), Putative antitoxin HigA3 | | Authors: | Park, J.Y, Lee, B.J. | | Deposit date: | 2020-01-23 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Induced DNA bending by unique dimerization of HigA antitoxin.
Iucrj, 7, 2020
|
|
6LTZ
 
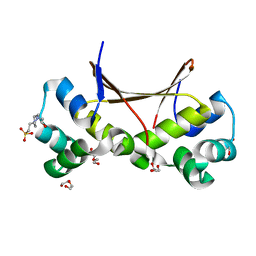 | |
1LLI
 
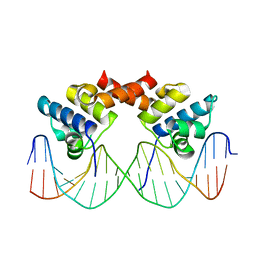 | | THE CRYSTAL STRUCTURE OF A MUTANT PROTEIN WITH ALTERED BUT IMPROVED HYDROPHOBIC CORE PACKING | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Lim, W.A, Hodel, A, Sauer, R.T, Richards, F.M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a mutant protein with altered but improved hydrophobic core packing.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
4OB4
 
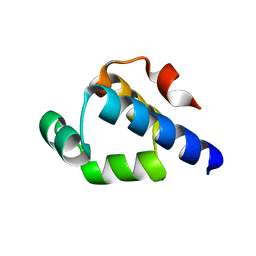 | | Structure of the S. venezulae BldD DNA-binding domain | | Descriptor: | Putative DNA-binding protein | | Authors: | schumacher, M.A, Tschowri, N, Buttner, M, Brennan, R. | | Deposit date: | 2014-01-06 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell(Cambridge,Mass.), 158, 2014
|
|
6RNZ
 
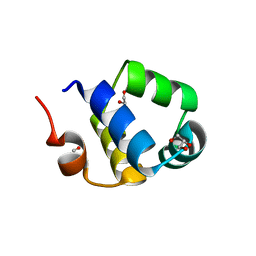 | | Crystal structure of the N-terminal HTH DNA-binding domain of the essential repressor DdrO from radiation-resistant Deinococcus bacteria (Deinococcus deserti) | | Descriptor: | GLYCEROL, HTH-type transcriptional regulator DdrOC | | Authors: | Arnoux, P, Siponen, M.I, Pignol, D, Brandelet, G, De Groot, A, Blanchard, L. | | Deposit date: | 2019-05-10 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the transcriptional repressor DdrO: insight into the metalloprotease/repressor-controlled radiation response in Deinococcus.
Nucleic Acids Res., 47, 2019
|
|
5WOQ
 
 | |
3S8Q
 
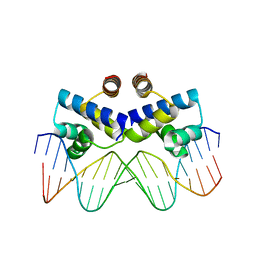 | | Crystal structure of the R-M controller protein C.Esp1396I OL operator complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*AP*CP*TP*TP*AP*TP*AP*GP*TP*CP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*GP*GP*AP*CP*TP*AP*TP*AP*AP*GP*TP*CP*AP*CP*A)-3'), R-M CONTROLLER PROTEIN | | Authors: | McGeehan, J.E, Ball, N.J, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | Deposit date: | 2011-05-30 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of dual symmetry by the controller protein C.Esp1396I based on the structure of the transcriptional activation complex.
Nucleic Acids Res., 40, 2012
|
|
7CSW
 
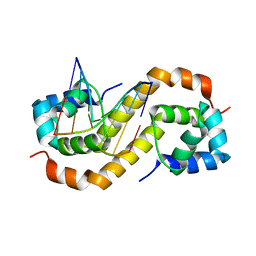 | | Pseudomonas aeruginosa antitoxin HigA with pa2440 promoter | | Descriptor: | HTH cro/C1-type domain-containing protein, pa2440 | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
7CSY
 
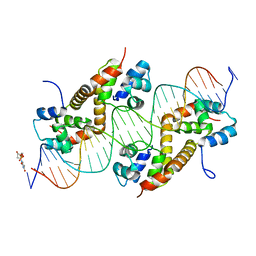 | | Pseudomonas aeruginosa antitoxin HigA with higBA promoter | | Descriptor: | DNA (28-MER), DNA (29-MER), HTH cro/C1-type domain-containing protein, ... | | Authors: | Song, Y.J, Luo, G.H, Bao, R. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Pseudomonas aeruginosa antitoxin HigA functions as a diverse regulatory factor by recognizing specific pseudopalindromic DNA motifs.
Environ.Microbiol., 23, 2021
|
|
3KZ3
 
 | |
3ZHI
 
 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 | | Descriptor: | CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
3KXA
 
 | | Crystal Structure of NGO0477 from Neisseria gonorrhoeae | | Descriptor: | ASPARAGINE, CHLORIDE ION, Putative uncharacterized protein, ... | | Authors: | Ren, J, Sainsbury, S, Nettleship, J.E, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-12-02 | | Release date: | 2010-01-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of NGO0477 from Neisseria gonorrhoeae reveals a novel protein fold incorporating a helix-turn-helix motif.
Proteins, 78, 2010
|
|
1X57
 
 | | Solution structures of the HTH domain of human EDF-1 protein | | Descriptor: | Endothelial differentiation-related factor 1 | | Authors: | Nameki, N, Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-15 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the HTH domain of human EDF-1 protein
To be Published
|
|
3LFP
 
 | |
3VK0
 
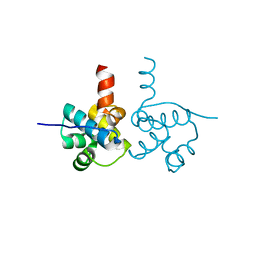 | | Crystal Structure of hypothetical transcription factor NHTF from Neisseria | | Descriptor: | Transcriptional regulator | | Authors: | Wang, H.-C, Ko, T.-P, Wu, M.-L, Wu, H.-J, Ku, S.-C, Wang, A.H.-J. | | Deposit date: | 2011-11-01 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Neisseria conserved protein DMP19 is a DNA mimic protein that prevents DNA binding to a hypothetical nitrogen-response transcription factor
Nucleic Acids Res., 2012
|
|
3LIS
 
 | |
4F8D
 
 | |
3UFD
 
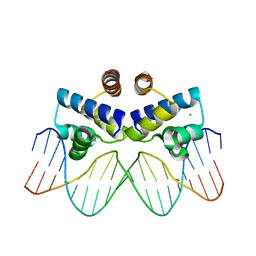 | | C.Esp1396I bound to its highest affinity operator site OM | | Descriptor: | CHLORIDE ION, DNA (5'-D(*AP*TP*GP*TP*AP*GP*AP*CP*TP*AP*TP*AP*GP*TP*CP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*GP*AP*CP*TP*AP*TP*AP*GP*TP*CP*TP*AP*CP*A)-3'), ... | | Authors: | Ball, N.J, McGeehan, J.E, Streeter, S.D, Thresh, S.-J, Kneale, G.G. | | Deposit date: | 2011-11-01 | | Release date: | 2012-07-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of differential DNA sequence recognition by restriction-modification controller proteins.
Nucleic Acids Res., 40, 2012
|
|
3BS3
 
 | | Crystal structure of a putative DNA-binding protein from Bacteroides fragilis | | Descriptor: | 1,2-ETHANEDIOL, Putative DNA-binding protein, SULFATE ION | | Authors: | Cuff, M.E, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of a putative DNA-binding protein from Bacteroides fragilis.
TO BE PUBLISHED
|
|
4FN3
 
 | |
4FBI
 
 | |
3ZHM
 
 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 in complex with the OL2 operator half-site | | Descriptor: | 5'-D(*AP*CP*GP*TP*GP*AP*AP*CP*TP*TP*GP*CP*AP*CP *TP*TP*GP*A)-3', 5'-D(*AP*GP*TP*TP*CP*AP*CP*GP*TP*TP*CP*AP*AP*GP *TP*GP*CP*A)-3', CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-22 | | Release date: | 2013-12-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
1UTX
 
 | | Regulation of Cytolysin Expression by Enterococcus faecalis: Role of CylR2 | | Descriptor: | CYLR2, IODIDE ION, SODIUM ION | | Authors: | Razeto, A, Rumpel, S, Pillar, C.M, Gilmore, M.S, Becker, S, Zweckstetter, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and DNA-Binding Properties of the Cytolysin Regulator CylR2 from Enterococcus Faecalis
Embo J., 23, 2004
|
|
