1DUC
 
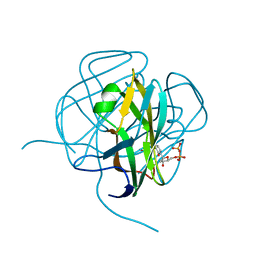 | | EIAV DUTPASE DUDP/STRONTIUM COMPLEX | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE, DEOXYURIDINE-5'-DIPHOSPHATE, STRONTIUM ION | | Authors: | Dauter, Z, Persson, R, Rosengren, A.M, Nyman, P.O, Wilson, K.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 1997-11-29 | | Release date: | 1998-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex.
J.Mol.Biol., 285, 1999
|
|
1DUD
 
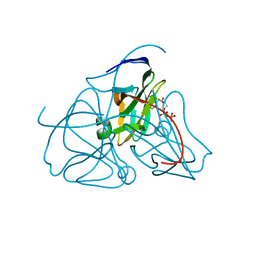 | |
1DUE
 
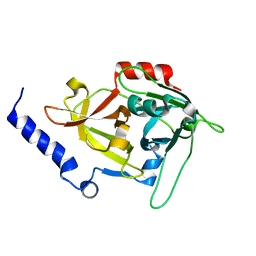 | |
1DUF
 
 | | THE NMR STRUCTURE OF DNA DODECAMER DETERMINED IN AQUEOUS DILUTE LIQUID CRYSTALLINE PHASE | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Tjandra, N, Tate, S, Ono, A, Kainosho, M, Bax, A. | | Deposit date: | 2000-01-17 | | Release date: | 2000-07-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR Structure of a DNA Dodecamer in an Aqueous Dilute Liquid Crystalline Phase
J.Am.Chem.Soc., 122, 2000
|
|
1DUG
 
 | | STRUCTURE OF THE FIBRINOGEN G CHAIN INTEGRIN BINDING AND FACTOR XIIIA CROSSLINKING SITES OBTAINED THROUGH CARRIER PROTEIN DRIVEN CRYSTALLIZATION | | Descriptor: | GLUTATHIONE, chimera of GLUTATHIONE S-TRANSFERASE-synthetic LINKEr-C-TERMINAL FIBRINOGEN GAMMA CHAIN | | Authors: | Ware, S, Donahue, J.P, Hawiger, J, Anderson, W.F. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the fibrinogen gamma-chain integrin binding and factor XIIIa cross-linking sites obtained through carrier protein driven crystallization.
Protein Sci., 8, 1999
|
|
1DUH
 
 | | CRYSTAL STRUCTURE OF THE CONSERVED DOMAIN IV OF E. COLI 4.5S RNA | | Descriptor: | 4.5S RNA DOMAIN IV, LUTETIUM (III) ION, MAGNESIUM ION, ... | | Authors: | Jovine, L, Hainzl, T, Oubridge, C, Scott, W.G, Li, J, Sixma, T.K, Wonacott, A, Skarzynski, T, Nagai, K. | | Deposit date: | 2000-01-17 | | Release date: | 2000-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA.
Structure Fold.Des., 8, 2000
|
|
1DUI
 
 | | Subtilisin BPN' from Bacillus amyloliquefaciens, crystal growth mutant | | Descriptor: | DIISOPROPYL PHOSPHONATE, PROTEIN (SUBTILISIN BPN'), SODIUM ION | | Authors: | Pan, Q, Gallagher, D.T. | | Deposit date: | 2000-01-17 | | Release date: | 2000-01-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing Protein Interaction Chemistry Through Crystal Growth: Structure, Mutation, and Mechanism in Subtilisin s88
J.Cryst.Growth, 212, 2000
|
|
1DUJ
 
 | | SOLUTION STRUCTURE OF THE SPINDLE ASSEMBLY CHECKPOINT PROTEIN HUMAN MAD2 | | Descriptor: | SPINDLE ASSEMBLY CHECKPOINT PROTEIN | | Authors: | Luo, X, Fang, G, Coldiron, M, Lin, Y, Yu, H. | | Deposit date: | 2000-01-17 | | Release date: | 2000-03-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20.
Nat.Struct.Biol., 7, 2000
|
|
1DUK
 
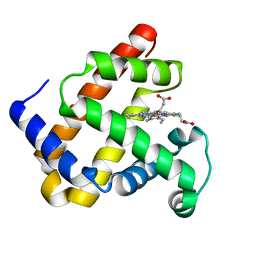 | | WILD-TYPE RECOMBINANT SPERM WHALE METAQUOMYOGLOBIN | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, WILD-TYPE RECOMBINANT SPERM WHALE METAQUOMYOGLOBIN | | Authors: | Barrick, D, Dahlquist, F.W. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Trans-substitution of the proximal hydrogen bond in myoglobin: I. Structural consequences of hydrogen bond deletion.
Proteins, 39, 2000
|
|
1DUL
 
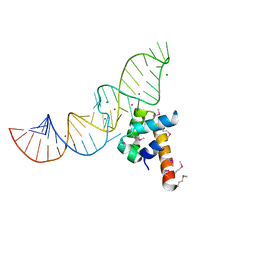 | | STRUCTURE OF THE RIBONUCLEOPROTEIN CORE OF THE E. COLI SIGNAL RECOGNITION PARTICLE | | Descriptor: | 4.5 S RNA DOMAIN IV, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Batey, R.T, Rambo, R.P, Lucast, L, Rha, B, Doudna, J.A. | | Deposit date: | 2000-01-17 | | Release date: | 2000-02-28 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ribonucleoprotein core of the signal recognition particle.
Science, 287, 2000
|
|
1DUM
 
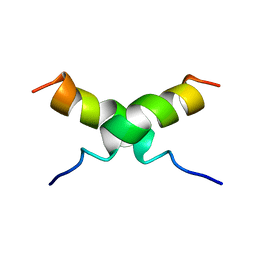 | | NMR STRUCTURE OF [F5Y, F16W] MAGAININ 2 BOUND TO PHOSPHOLIPID VESICLES | | Descriptor: | MAGAININ 2 | | Authors: | Takeda, A, Wakamatsu, K, Tachi, T, Matsuzaki, K. | | Deposit date: | 2000-01-18 | | Release date: | 2001-06-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Effects of peptide dimerization on pore formation: Antiparallel disulfide-dimerized magainin 2 analogue.
Biopolymers, 58, 2001
|
|
1DUN
 
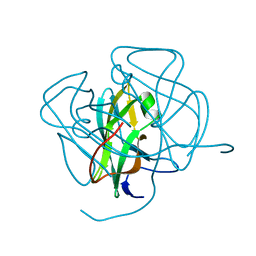 | | EIAV DUTPASE NATIVE | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Dauter, Z, Persson, R, Rosengren, A.M, Nyman, P.O, Wilson, K.S, Cedergren-Zeppezauer, E.S. | | Deposit date: | 1997-11-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex.
J.Mol.Biol., 285, 1999
|
|
1DUO
 
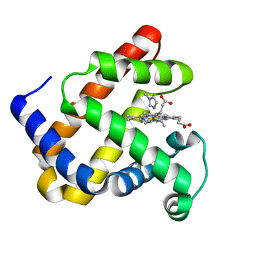 | |
1DUP
 
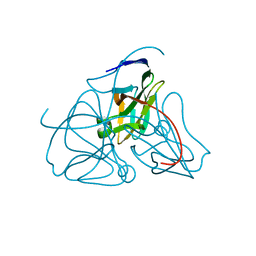 | | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDO HYDROLASE (D-UTPASE) | | Descriptor: | DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Dauter, Z, Wilson, K.S, Larsson, G, Nyman, P.O, Cedergren, E. | | Deposit date: | 1995-09-01 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a dUTPase.
Nature, 355, 1992
|
|
1DUQ
 
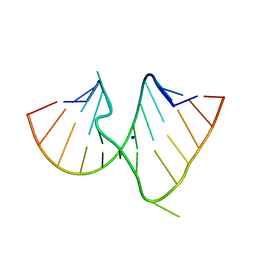 | |
1DUR
 
 | |
1DUS
 
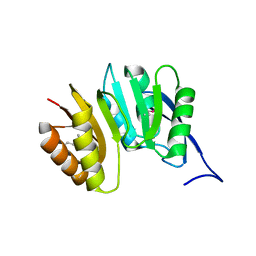 | | MJ0882-A hypothetical protein from M. jannaschii | | Descriptor: | MJ0882 | | Authors: | Hung, L, Huang, L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2000-01-18 | | Release date: | 2000-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based experimental confirmation of biochemical function to a methyltransferase, MJ0882, from hyperthermophile Methanococcus jannaschii
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
1DUT
 
 | | FIV DUTP PYROPHOSPHATASE | | Descriptor: | DUTP PYROPHOSPHATASE, MAGNESIUM ION | | Authors: | Prasad, G.S, Stura, E.A, Mcree, D.E, Laco, G.S, Hasselkus-Light, C, Elder, J.H, Stout, C.D. | | Deposit date: | 1996-09-15 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dUTP pyrophosphatase from feline immunodeficiency virus.
Protein Sci., 5, 1996
|
|
1DUV
 
 | | CRYSTAL STRUCTURE OF E. COLI ORNITHINE TRANSCARBAMOYLASE COMPLEXED WITH NDELTA-L-ORNITHINE-DIAMINOPHOSPHINYL-N-SULPHONIC ACID (PSORN) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NDELTA-(N'-SULPHODIAMINOPHOSPHINYL)-L-ORNITHINE, ORNITHINE TRANSCARBAMOYLASE | | Authors: | Langley, D.B, Templeton, M.D, Fields, B.A, Mitchell, R.E, Collyer, C.A. | | Deposit date: | 2000-01-18 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of inactivation of ornithine transcarbamoylase by Ndelta -(N'-Sulfodiaminophosphinyl)-L-ornithine, a true transition state analogue? Crystal structure and implications for catalytic mechanism.
J.Biol.Chem., 275, 2000
|
|
1DUW
 
 | | STRUCTURE OF NONAHEME CYTOCHROME C | | Descriptor: | GLYCEROL, HEME C, NONAHEME CYTOCHROME C | | Authors: | Umhau, S, Fritz, G, Diederichs, K, Breed, J, Kroneck, P.M, Welte, W. | | Deposit date: | 2000-01-19 | | Release date: | 2001-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Three-dimensional structure of the nonaheme cytochrome c from Desulfovibrio desulfuricans Essex in the Fe(III) state at 1.89 A resolution.
Biochemistry, 40, 2001
|
|
1DUX
 
 | | ELK-1/DNA STRUCTURE REVEALS HOW RESIDUES DISTAL FROM DNA-BINDING SURFACE AFFECT DNA-RECOGNITION | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*T)-3'), ETS-DOMAIN PROTEIN ELK-1 | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 2000-01-19 | | Release date: | 2000-04-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the elk-1-DNA complex reveals how DNA-distal residues affect ETS domain recognition of DNA.
Nat.Struct.Biol., 7, 2000
|
|
1DUY
 
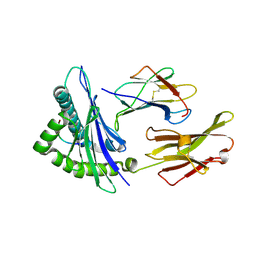 | | CRYSTAL STRUCTURE OF HLA-A*0201/OCTAMERIC TAX PEPTIDE COMPLEX | | Descriptor: | BETA-2 MICROGLOBULIN, HLA-A2*0201, HTLV-1 OCTAMERIC TAX PEPTIDE | | Authors: | Khan, A.R, Baker, B.M, Ghosh, P, Biddison, W.E, Wiley, D.C. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site.
J.Immunol., 164, 2000
|
|
1DUZ
 
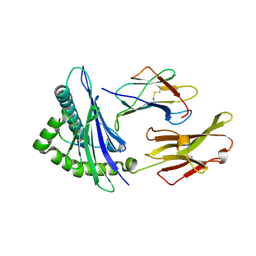 | | HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A 0201) IN COMPLEX WITH A NONAMERIC PEPTIDE FROM HTLV-1 TAX PROTEIN | | Descriptor: | BETA-2 MICROGLOBULIN, HLA-A*0201, HTLV-1 OCTAMERIC TAX PEPTIDE | | Authors: | Khan, A.R, Baker, B.M, Ghosh, P, Biddison, W.E, Wiley, D.C. | | Deposit date: | 2000-01-19 | | Release date: | 2000-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site.
J.Immunol., 164, 2000
|
|
1DV0
 
 | |
1DV1
 
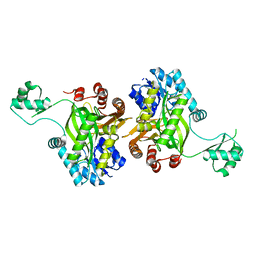 | | STRUCTURE OF BIOTIN CARBOXYLASE (APO) | | Descriptor: | BIOTIN CARBOXYLASE, PHOSPHATE ION | | Authors: | Thoden, J.B, Blanchard, C.Z, Holden, H.M, Waldrop, G.L. | | Deposit date: | 2000-01-19 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Movement of the biotin carboxylase B-domain as a result of ATP binding.
J.Biol.Chem., 275, 2000
|
|
