1X4T
 
 | | Solution structure of Isy1 domain in hypothetical protein | | Descriptor: | hypothetical protein LOC57905 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Isy1 domain in hypothetical protein
To be Published
|
|
1MQM
 
 | | BHA/LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | ha, y, steven, d.j, skehel, j.j, wiley, d.c. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus.
Virology, 309, 2003
|
|
1X5O
 
 | | Solution structure of RRM domain in RNA binding motif, single-stranded interacting protein 1 | | Descriptor: | RNA binding motif, single-stranded interacting protein 1 | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in RNA binding motif, single-stranded interacting protein 1
To be Published
|
|
1ZS7
 
 | | The structure of gene product APE0525 from Aeropyrum pernix | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, hypothetical protein APE0525 | | Authors: | Cuff, M.E, Skarina, T, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-23 | | Release date: | 2005-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of gene product APE0525 from Aeropyrum pernix
To be Published
|
|
1X23
 
 | | Crystal structure of ubch5c | | Descriptor: | Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Nakanishi, M, Teshima, N, Mizushima, T, Murata, S, Tanaka, K, Yamane, T. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of ubch5c
To be Published
|
|
1ZSR
 
 | | Crystal structure of wild type HIV-1 protease (BRU isolate) with a hydroxyethylamine peptidomimetic inhibitor BOC-PHE-PSI[S-CH(OH)CH2NH]-PHE-GLU-PHE-NH2 | | Descriptor: | N-{(2S,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-alpha-glutamyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-24 | | Release date: | 2006-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | On the role of the R configuration of the reaction-intermediate isostere in HIV-1 protease-inhibitor binding: X-ray structure at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1WG6
 
 | | Solution structure of PDZ domain in protein XP_110852 | | Descriptor: | HYPOTHETICAL PROTEIN (RIKEN cDNA 2810455B10) | | Authors: | He, F, Hanaki, K, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-27 | | Release date: | 2004-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PDZ domain in protein XP_110852
To be Published
|
|
1WHQ
 
 | | Solution structure of the N-terminal dsRBD from hypothetical protein BAB28848 | | Descriptor: | RNA helicase A | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal dsRBD from hypothetical protein BAB28848
To be Published
|
|
1MAG
 
 | |
1WJL
 
 | | Solution structure of PDZ domain of mouse Cypher protein | | Descriptor: | Cypher protein | | Authors: | Sato, M, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of PDZ domain of mouse Cypher protein
To be Published
|
|
1WJW
 
 | | Solution structure of the C-terminal domain of mouse phosphoacetylglucosamine mutase (PAGM) | | Descriptor: | Phosphoacetylglucosamine mutase | | Authors: | Yoneyama, M, Tochio, N, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of mouse phosphoacetylglucosamine mutase (PAGM)
To be Published
|
|
1ZZ0
 
 | | Crystal structure of a HDAC-like protein with acetate bound | | Descriptor: | ACETATE ION, Histone deacetylase-like amidohydrolase, POTASSIUM ION, ... | | Authors: | Nielsen, T.K, Hildmann, C, Dickmanns, A, Schwienhorst, A, Ficner, R. | | Deposit date: | 2005-06-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a bacterial class 2 histone deacetylase homologue
J.Mol.Biol., 354, 2005
|
|
220D
 
 | | INFLUENCE OF COUNTER-IONS ON THE CRYSTAL STRUCTURES OF DNA DECAMERS: BINDING OF [CO(NH3)6]3+ AND BA2+ TO A-DNA | | Descriptor: | BARIUM ION, DNA (5'-D(*AP*CP*CP*CP*GP*CP*GP*GP*GP*T)-3') | | Authors: | Gao, Y.-G, Robinson, H, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1995-06-26 | | Release date: | 1996-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influence of counter-ions on the crystal structures of DNA decamers: binding of [Co(NH3)6]3+ and Ba2+ to A-DNA.
Biophys.J., 69, 1995
|
|
223D
 
 | | DIRECT OBSERVATION OF TWO BASE-PAIRING MODES OF A CYTOSINE-THYMINE ANALOGUE WITH GUANINE IN A DNA Z-FORM DUPLEX: SIGNIFICANCE FOR BASE ANALOGUE MUTAGENESIS | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C46)P*G)-3') | | Authors: | Moore, M.H, Van Meervelt, L, Salisbury, S.A, Kong Thoo Lin, P, Brown, D.M. | | Deposit date: | 1995-08-01 | | Release date: | 1995-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Direct observation of two base-pairing modes of a cytosine-thymine analogue with guanine in a DNA Z-form duplex: significance for base analogue mutagenesis.
J.Mol.Biol., 251, 1995
|
|
228L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
1MEO
 
 | | human glycinamide ribonucleotide Transformylase at pH 4.2 | | Descriptor: | PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase, SULFATE ION | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
1MIC
 
 | |
1ZVO
 
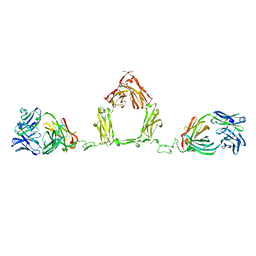 | | Semi-extended solution structure of human myeloma immunoglobulin D determined by constrained X-ray scattering | | Descriptor: | Immunoglobulin delta heavy chain, myeloma immunoglobulin D lambda | | Authors: | Sun, Z, Almogren, A, Furtado, P.B, Chowdhury, B, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2005-06-02 | | Release date: | 2005-10-25 | | Last modified: | 2024-02-14 | | Method: | SOLUTION SCATTERING | | Cite: | Semi-extended Solution Structure of Human Myeloma Immunoglobulin D Determined by Constrained X-ray Scattering.
J.Mol.Biol., 353, 2005
|
|
1X5P
 
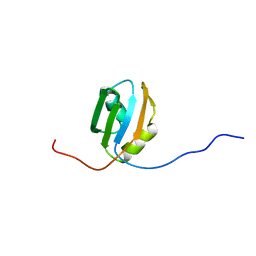 | | Solution structure of RRM domain in Parp14 | | Descriptor: | Negative elongation factor E | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in Parp14
To be Published
|
|
1MP2
 
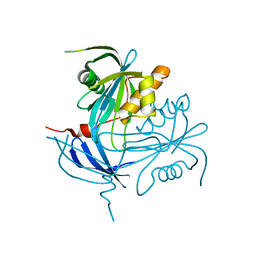 | | Structure of MT-ADPRase (Apoenzyme), a Nudix hydrolase from Mycobacterium tuberculosis | | Descriptor: | ADPR pyrophosphatase | | Authors: | Kang, L.-W, Gabelli, S.B, Bianchet, M.A, Cunningham, J.E, O'Handley, S.F, Amzel, L.M. | | Deposit date: | 2002-09-11 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of MT-ADPRase, a Nudix hydrolase from Mycobacterium tuberculosis
Structure, 11, 2003
|
|
221D
 
 | |
231L
 
 | | T4 LYSOZYME MUTANT M106K | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-03 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
1X4D
 
 | | Solution structure of RRM domain in Matrin 3 | | Descriptor: | Matrin 3 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RRM domain in Matrin 3
To be Published
|
|
255D
 
 | |
1MQN
 
 | | BHA/LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1 chain, ... | | Authors: | ha, y, stevens, d.j, skehel, j.j, wiley, d.c. | | Deposit date: | 2002-09-16 | | Release date: | 2003-08-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus.
Virology, 309, 2003
|
|
