8FJ5
 
 | | Structure of the Haloferax volcanii archaeal type IV pilus | | Descriptor: | Pilin_N domain-containing protein | | Authors: | Wang, F, Kreutzberger, M.A, Baquero, D.P, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-12-19 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FJS
 
 | |
6NM5
 
 | | F-pilus/MS2 Maturation protein complex | | Descriptor: | (2R)-2,3-dihydroxypropyl ethyl hydrogen (S)-phosphate, Maturation protein, Type IV conjugative transfer system pilin TraA | | Authors: | Meng, R, Chang, J, Zhang, J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural basis for the adsorption of a single-stranded RNA bacteriophage.
Nat Commun, 10, 2019
|
|
6T8S
 
 | |
5Y9H
 
 | | Crystal structure of SafDAA-dsc complex | | Descriptor: | SafD,Putative outer membrane protein,Putative outer membrane protein,Putative outer membrane protein | | Authors: | Zeng, L.H, Zhang, L, Wang, P.R, Meng, G. | | Deposit date: | 2017-08-25 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of host recognition and biofilm formation by Salmonella Saf pili
Elife, 6, 2017
|
|
6VK9
 
 | | Cryo-EM structure of PilA-N/C from Geobacter sulfurreducens | | Descriptor: | Geopilin domain 1 protein, Geopilin domain 2 protein | | Authors: | Gu, Y, Srikanth, V, Malvankar, N.S, Samatey, F.A. | | Deposit date: | 2020-01-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Geobacter pili reveals secretory rather than nanowire behaviour
Nature, 597, 2021
|
|
1QPP
 
 | |
1QPX
 
 | |
1L4I
 
 | | Crystal Structure of the Periplasmic Chaperone SfaE | | Descriptor: | SfaE PROTEIN | | Authors: | Knight, S.D, Choudhury, D, Hultgren, S, Pinkner, J, Stojanoff, V, Thompson, A. | | Deposit date: | 2002-03-05 | | Release date: | 2002-06-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the S pilus periplasmic chaperone SfaE at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3JVU
 
 | |
6CD2
 
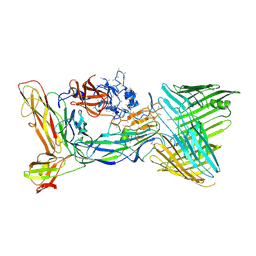 | | Crystal structure of the PapC usher bound to the chaperone-adhesin PapD-PapG | | Descriptor: | Chaperone protein PapD, Outer membrane usher protein PapC, PapGII adhesin protein | | Authors: | Omattage, N.S, Deng, Z, Yuan, P, Hultgren, S.J. | | Deposit date: | 2018-02-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for usher activation and intramolecular subunit transfer in P pilus biogenesis in Escherichia coli.
Nat Microbiol, 3, 2018
|
|
4QS4
 
 | |
6FJY
 
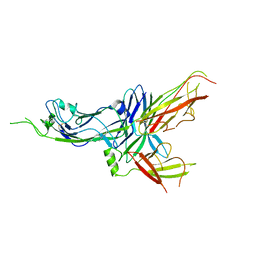 | | Crystal structure of CsuC-CsuE chaperone-tip adhesion subunit pre-assembly complex from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuC, Protein CsuE | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-16 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis forAcinetobacter baumanniibiofilm formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3JVV
 
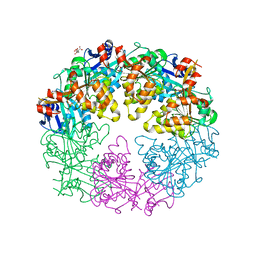 | | Crystal Structure of P. aeruginosa PilT with bound AMP-PCP | | Descriptor: | CITRIC ACID, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Misic, A.M, Satyshur, K.A, Forest, K.T. | | Deposit date: | 2009-09-17 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | P. aeruginosa PilT structures with and without nucleotide reveal a dynamic type IV pilus retraction motor.
J.Mol.Biol., 400, 2010
|
|
5K93
 
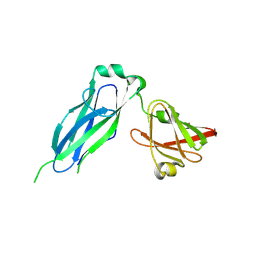 | | PapD wild-type chaperone | | Descriptor: | Chaperone protein PapD | | Authors: | Sarowar, S, Li, H. | | Deposit date: | 2016-05-31 | | Release date: | 2016-07-20 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Escherichia coli P and Type 1 Pilus Assembly Chaperones PapD and FimC Are Monomeric in Solution.
J.Bacteriol., 198, 2016
|
|
6R74
 
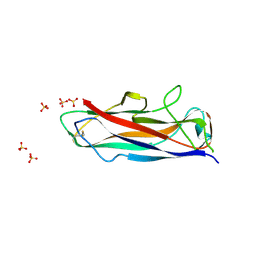 | | N-terminally reversed variant of FimA E. coli | | Descriptor: | SULFATE ION, Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Echeverria, B, Glockshuber, R. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Donor strand sequence, rather than donor strand orientation, determines the stability and non-equilibrium folding of the type 1 pilus subunit FimA.
J.Biol.Chem., 295, 2020
|
|
7CBS
 
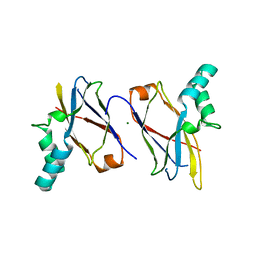 | | Crystal structure of SpaB basal pilin from Lactobacillus rhamnosus GG | | Descriptor: | CHLORIDE ION, LPXTG cell wall anchor domain-containing protein, MAGNESIUM ION | | Authors: | Megta, A.K, Pratap, S, Kant, A, Krishnan, V. | | Deposit date: | 2020-06-13 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the atypically adhesive SpaB basal pilus subunit: Mechanistic insights about its incorporation in lactobacillar SpaCBA pili.
Curr Res Struct Biol, 2, 2020
|
|
5UTT
 
 | | SrtA sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Sortase | | Authors: | Osipiuk, J, Ma, X, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cell-to-cell interaction requires optimal positioning of a pilus tip adhesin modulated by gram-positive transpeptidase enzymes.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3RCC
 
 | |
6R7E
 
 | | N-terminally reversed variant of FimA E. coli with alanine insertion at position 20 | | Descriptor: | FimA, SULFATE ION | | Authors: | Zyla, D, Echeverria, B, Glockshuber, R. | | Deposit date: | 2019-03-28 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Donor strand sequence, rather than donor strand orientation, determines the stability and non-equilibrium folding of the type 1 pilus subunit FimA.
J.Biol.Chem., 295, 2020
|
|
6S09
 
 | | C-terminally extended and N-terminally truncated variant of FimA E. coli at 1.5 Angstrom resolution | | Descriptor: | ACETIC ACID, FimA, SODIUM ION, ... | | Authors: | Zyla, D, Echeverria, B, Glockshuber, R. | | Deposit date: | 2019-06-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Donor strand sequence, rather than donor strand orientation, determines the stability and non-equilibrium folding of the type 1 pilus subunit FimA.
J.Biol.Chem., 295, 2020
|
|
6SWH
 
 | | Crystal structure of the ternary complex between the type 1 pilus proteins FimC, FimI and FimA from E. coli | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Giese, C, Puorger, C, Ignatov, O, Weber, M, Scharer, M.A, Capitani, G, Glockshuber, R. | | Deposit date: | 2019-09-20 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
5NCA
 
 | | Solution structure of ComGC from Streptococcus pneumoniae | | Descriptor: | Competence protein ComGC | | Authors: | Erlendsson, S, Schmeider, P, Lichtenberg, C, Teilum, K, Akbey, U. | | Deposit date: | 2017-03-03 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the competence pilus major pilin ComGC in Streptococcus pneumoniae.
J. Biol. Chem., 292, 2017
|
|
7B0X
 
 | | Crystal structure of the ternary complex of the E. coli type 1 pilus proteins FimC, FimI and the N-terminal domain of FimD | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein FimC, Fimbrin-like protein FimI, ... | | Authors: | Scharer, M.A, Zigova, Z, Giese, C, Puorger, C, Ignatov, O, Capitani, G, Glockshuber, R. | | Deposit date: | 2020-11-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Comprehensive kinetic characterization of bacterial pilus rod assembly and assembly termination
To Be Published
|
|
3RBK
 
 | |
