1D9O
 
 | |
1D9P
 
 | |
1D9Q
 
 | | OXIDIZED PEA FRUCTOSE-1,6-BISPHOSPHATASE FORM 1 | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE | | Authors: | Chiadmi, M, Navaza, A, Miginiac-Maslow, M, Jacquot, J.-P, Cherfils, J. | | Deposit date: | 1999-10-29 | | Release date: | 1999-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Redox signalling in the chloroplast: structure of oxidized pea fructose-1,6-bisphosphate phosphatase.
EMBO J., 18, 1999
|
|
1D9R
 
 | | CRYSTAL STRUCTURE OF DNA SHEARED TANDEM G-A BASE PAIRS | | Descriptor: | 5'-D(*CP*CP*GP*AP*AP*(BRU)P*GP*AP*GP*G)-3', COBALT HEXAMMINE(III) | | Authors: | Gao, Y.-G, Robinson, H, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 1999-10-29 | | Release date: | 1999-11-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and recognition of sheared tandem G x A base pairs associated with human centromere DNA sequence at atomic resolution.
Biochemistry, 38, 1999
|
|
1D9S
 
 | | TUMOR SUPPRESSOR P15(INK4B) STRUCTURE BY COMPARATIVE MODELING AND NMR DATA | | Descriptor: | CYCLIN-DEPENDENT KINASE 4 INHIBITOR B | | Authors: | Yuan, C, Ji, L, Selby, T.L, Byeon, I.J.L, Tsai, M.D. | | Deposit date: | 1999-10-29 | | Release date: | 2000-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor INK4: comparisons of conformational properties between p16(INK4A) and p18(INK4C).
J.Mol.Biol., 294, 1999
|
|
1D9U
 
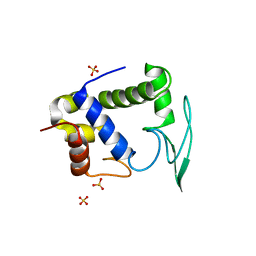 | | BACTERIOPHAGE LAMBDA LYSOZYME COMPLEXED WITH A CHITOHEXASACHARIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BACTERIOPHAGE LAMBDA LYSOZYME, ... | | Authors: | Leung, A.K.W, Duewel, H.S, Honek, J.F, Berghuis, A.M. | | Deposit date: | 1999-10-30 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the lytic transglycosylase from bacteriophage lambda in complex with hexa-N-acetylchitohexaose.
Biochemistry, 40, 2001
|
|
1D9V
 
 | |
1D9W
 
 | |
1D9X
 
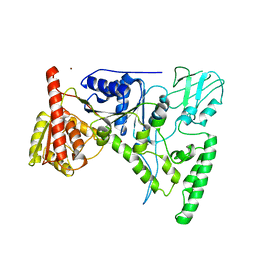 | | CRYSTAL STRUCTURE OF THE DNA REPAIR PROTEIN UVRB | | Descriptor: | EXCINUCLEASE UVRABC COMPONENT UVRB, ZINC ION | | Authors: | Theis, K, Chen, P.J, Skorvaga, M, Van Houten, B, Kisker, C. | | Deposit date: | 1999-10-30 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.
EMBO J., 18, 1999
|
|
1D9Y
 
 | | NEISSERIA GONORRHOEAE FERRIC BINDING PROTEIN | | Descriptor: | FE (III) ION, PHOSPHATE ION, PROTEIN (PERIPLASMIC IRON-BINDING PROTEIN) | | Authors: | McRee, D.E, Bruns, C.M, Williams, P.A, Mietzner, T.A, Nunn, R. | | Deposit date: | 1999-10-30 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Iron Uptake in the Pathogen Neisseria gonorrhoeae
To be Published
|
|
1D9Z
 
 | | CRYSTAL STRUCTURE OF THE DNA REPAIR PROTEIN UVRB IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, EXCINUCLEASE UVRABC COMPONENT UVRB, MAGNESIUM ION, ... | | Authors: | Theis, K, Chen, P.J, Skorvaga, M, Van Houten, B, Kisker, C. | | Deposit date: | 1999-10-30 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair.
EMBO J., 18, 1999
|
|
1DA0
 
 | | DNA-DRUG INTERACTIONS: THE CRYSTAL STRUCTURE OF D(CGATCG) COMPLEXED WITH DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Moore, M.H, Hunter, W.N, Langlois D'Estaintot, B, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA-drug interactions. The crystal structure of d(CGATCG) complexed with daunomycin.
J.Mol.Biol., 206, 1989
|
|
1DA1
 
 | | STRUCTURAL CHARACTERISATION OF THE BROMOURACIL-GUANINE BASE PAIR MISMATCH IN A Z-DNA FRAGMENT | | Descriptor: | DNA (5'-D(*(BRU)P*GP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Brown, T, Kneale, G, Hunter, W.N, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural characterisation of the bromouracil.guanine base pair mismatch in a Z-DNA fragment.
Nucleic Acids Res., 14, 1986
|
|
1DA2
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO4CG): N4-METHOXYCYTOSINE/GUANINE BASE-PAIRS IN Z-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(C45)P*G)-3') | | Authors: | Van Meervelt, L, Moore, M.H, Lin, P.K.T, Brown, D.M, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular and crystal structure of d(CGCGmo4CG): N4-methoxycytosine.guanine base-pairs in Z-DNA.
J.Mol.Biol., 216, 1990
|
|
1DA3
 
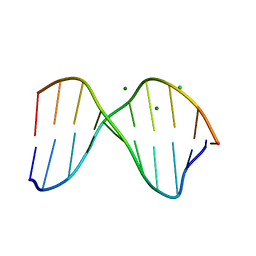 | | THE CRYSTAL STRUCTURE OF THE TRIGONAL DECAMER C-G-A-T-C-G-6MEA-T-C-G: A B-DNA HELIX WITH 10.6 BASE-PAIRS PER TURN | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*AP*TP*CP*GP*(6MA)P*TP*CP*G)-3'), MAGNESIUM ION | | Authors: | Baikalov, I, Grzeskowiak, K, Yanagi, K, Quintana, J, Dickerson, R.E. | | Deposit date: | 1992-11-09 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the trigonal decamer C-G-A-T-C-G-6meA-T-C-G: a B-DNA helix with 10.6 base-pairs per turn.
J.Mol.Biol., 231, 1993
|
|
1DA4
 
 | |
1DA5
 
 | |
1DA9
 
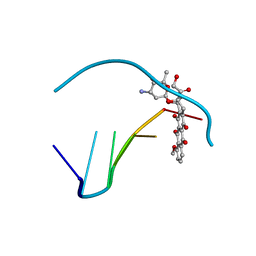 | | ANTHRACYCLINE-DNA INTERACTIONS AT UNFAVOURABLE BASE BASE-PAIR TRIPLET-BINDING SITES: STRUCTURES OF D(CGGCCG)/DAUNOMYCIN AND D(TGGCCA)/ADRIAMYCIN COMPL | | Descriptor: | DNA (5'-D(*TP*GP*GP*CP*CP*A)-3'), DOXORUBICIN | | Authors: | Leonard, G.A, Hambley, T.W, McAuley-Hecht, K, Brown, T, Hunter, W.N. | | Deposit date: | 1993-01-21 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anthracycline-DNA interactions at unfavourable base-pair triplet-binding sites: structures of d(CGGCCG)/daunomycin and d(TGGCCA)/adriamycin complexes.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1DAA
 
 | |
1DAB
 
 | |
1DAD
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1DAE
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 3-(1-AMINOETHYL) NONANEDIOIC ACID | | Descriptor: | 3-(1-AMINOETHYL)NONANEDIOIC ACID, DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1DAF
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, ADP, AND CALCIUM | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1DAG
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID AND 5'-ADENOSYL-METHYLENE-TRIPHOSPHATE | | Descriptor: | 7-(CARBOXYAMINO)-8-AMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
1DAH
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 7,8-DIAMINO-NONANOIC ACID, 5'-ADENOSYL-METHYLENE-TRIPHOSPHATE, AND MANGANESE | | Descriptor: | 7,8-DIAMINO-NONANOIC ACID, DETHIOBIOTIN SYNTHETASE, MANGANESE (II) ION, ... | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
