1C7N
 
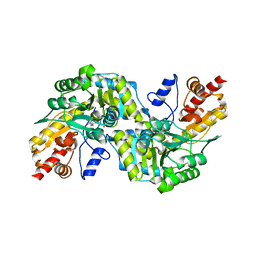 | | CRYSTAL STRUCTURE OF CYSTALYSIN FROM TREPONEMA DENTICOLA CONTAINS A PYRIDOXAL 5'-PHOSPHATE COFACTOR | | Descriptor: | CYSTALYSIN, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Krupka, H.I, Huber, R, Holt, S.C, Clausen, T. | | Deposit date: | 2000-03-16 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5'-phosphate-dependent protein acting as a haemolytic enzyme.
EMBO J., 19, 2000
|
|
1C9C
 
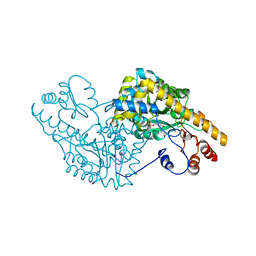 | | ASPARTATE AMINOTRANSFERASE COMPLEXED WITH C3-PYRIDOXAL-5'-PHOSPHATE | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1C7O
 
 | | CRYSTAL STRUCTURE OF CYSTALYSIN FROM TREPONEMA DENTICOLA CONTAINS A PYRIDOXAL 5'-PHOSPHATE-L-AMINOETHOXYVINYLGLYCINE COMPLEX | | Descriptor: | (2E,3E)-4-(2-aminoethoxy)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, CYSTALYSIN | | Authors: | Krupka, H.I, Huber, R, Holt, S.C, Clausen, T. | | Deposit date: | 2000-03-16 | | Release date: | 2000-07-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5'-phosphate-dependent protein acting as a haemolytic enzyme.
EMBO J., 19, 2000
|
|
1CQ7
 
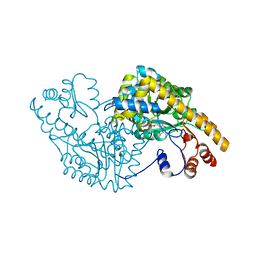 | | ASPARTATE AMINOTRANSFERASE (E.C. 2.6.1.1) COMPLEXED WITH C5-PYRIDOXAL-5P-PHOSPHATE | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO-PENTANOIC ACID, ASPARTATE AMINOTRANSFERASE | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-06 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1BJW
 
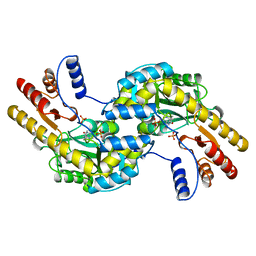 | | ASPARTATE AMINOTRANSFERASE FROM THERMUS THERMOPHILUS | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PHOSPHATE ION | | Authors: | Nakai, T, Okada, K, Kuramitsu, S, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-06-30 | | Release date: | 1999-07-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate.
Biochemistry, 38, 1999
|
|
1CQ6
 
 | | ASPARTATE AMINOTRANSFERASE COMPLEX WITH C4-PYRIDOXAL-5P-PHOSPHATE | | Descriptor: | 2-[O-PHOSPHONOPYRIDOXYL]-AMINO- BUTYRIC ACID, ASPARTATE AMINOTRANSFERASE | | Authors: | Ishijima, J, Nakai, T, Kawaguchi, S, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 1999-08-06 | | Release date: | 2000-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Free energy requirement for domain movement of an enzyme
J.Biol.Chem., 275, 2000
|
|
1DJU
 
 | | CRYSTAL STRUCTURE OF AROMATIC AMINOTRANSFERASE FROM PYROCOCCUS HORIKOSHII OT3 | | Descriptor: | AROMATIC AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matsui, I, Matsui, E, Sakai, Y, Kikuchi, H, Kawarabayashi, H. | | Deposit date: | 1999-12-06 | | Release date: | 2001-04-11 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular structure of hyperthermostable aromatic aminotransferase with novel substrate specificity from Pyrococcus horikoshii.
J.Biol.Chem., 275, 2000
|
|
1FG7
 
 | |
1FC4
 
 | | 2-AMINO-3-KETOBUTYRATE COA LIGASE | | Descriptor: | 2-AMINO-3-KETOBUTYRATE CONENZYME A LIGASE, 2-AMINO-3-KETOBUTYRIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Schmidt, A, Matte, A, Li, Y, Sivaraman, J, Larocque, R, Schrag, J.D, Smith, C, Sauve, V, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2000-07-17 | | Release date: | 2001-05-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of 2-amino-3-ketobutyrate CoA ligase from Escherichia coli complexed with a PLP-substrate intermediate: inferred reaction mechanism.
Biochemistry, 40, 2001
|
|
1FG3
 
 | | CRYSTAL STRUCTURE OF L-HISTIDINOL PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH L-HISTIDINOL | | Descriptor: | HISTIDINOL PHOSPHATE AMINOTRANSFERASE, PHOSPHORIC ACID MONO-[2-AMINO-3-(3H-IMIDAZOL-4-YL)-PROPYL]ESTER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Sivaraman, J, Cygler, M. | | Deposit date: | 2000-07-27 | | Release date: | 2001-08-22 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of histidinol phosphate aminotransferase (HisC) from Escherichia coli, and its covalent complex with pyridoxal-5'-phosphate and l-histidinol phosphate.
J.Mol.Biol., 311, 2001
|
|
1DJE
 
 | | CRYSTAL STRUCTURE OF THE PLP-BOUND FORM OF 8-AMINO-7-OXONANOATE SYNTHASE | | Descriptor: | 8-AMINO-7-OXONANOATE SYNTHASE, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION | | Authors: | Webster, S.P, Alexeev, D, Campopiano, D.J, Watt, R.M, Alexeeva, M, Sawyer, L, Baxter, R.L. | | Deposit date: | 1999-12-02 | | Release date: | 2000-12-04 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Mechanism of 8-amino-7-oxononanoate synthase: spectroscopic, kinetic, and crystallographic studies.
Biochemistry, 39, 2000
|
|
1GD9
 
 | | CRYSTALL STRUCTURE OF PYROCOCCUS PROTEIN-A1 | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-22 | | Release date: | 2001-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
1G7X
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R292L/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1BKG
 
 | | ASPARTATE AMINOTRANSFERASE FROM THERMUS THERMOPHILUS WITH MALEATE | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, MALEIC ACID | | Authors: | Nakai, T, Okada, K, Kuramitsu, S, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1998-07-07 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermus thermophilus HB8 aspartate aminotransferase and its complex with maleate.
Biochemistry, 38, 1999
|
|
1GC4
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 2 COMPLEXED WITH ASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-07-19 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
1GEY
 
 | | CRYSTAL STRUCTURE OF HISTIDINOL-PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH N-(5'-PHOSPHOPYRIDOXYL)-L-GLUTAMATE | | Descriptor: | 4-[(1,3-DICARBOXY-PROPYLAMINO)-METHYL]-3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDINIUM, HISTIDINOL-PHOSPHATE AMINOTRANSFERASE | | Authors: | Haruyama, K, Nakai, T, Miyahara, I, Hirotsu, K, Mizuguchi, H, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1G4X
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R292L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H. | | Deposit date: | 2000-10-29 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1G4V
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/Y225F | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-10-28 | | Release date: | 2000-11-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1GC3
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE TETRA MUTANT 2 COMPLEXED WITH TRYPTOPHAN | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-07-18 | | Release date: | 2001-09-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
1GEW
 
 | | CRYSTAL STRUCTURE OF HISTIDINOL-PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH PYRIDOXAL 5'-PHOSPHATE | | Descriptor: | HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Haruyama, K, Nakai, T, Miyahara, I, Hirotsu, K, Mizuguchi, H, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1G7W
 
 | | ASPARTATE AMINOTRANSFERASE ACTIVE SITE MUTANT N194A/R386L | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Mizuguchi, H, Hayashi, H, Okada, K, Miyahara, I, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2000-11-15 | | Release date: | 2000-11-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strain is more important than electrostatic interaction in controlling the pKa of the catalytic group in aspartate aminotransferase.
Biochemistry, 40, 2001
|
|
1GCK
 
 | | THERMUS THERMOPHILUS ASPARTATE AMINOTRANSFERASE DOUBLE MUTANT 1 COMPLEXED WITH ASPARTATE | | Descriptor: | ASPARTATE AMINOTRANSFERASE, ASPARTIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Nakai, T, Hirotsu, K, Kuramitsu, S. | | Deposit date: | 2000-08-04 | | Release date: | 2001-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate recognition mechanism of thermophilic dual-substrate enzyme.
J.Biochem., 130, 2001
|
|
1GEX
 
 | | CRYSTAL STRUCTURE OF HISTIDINOL-PHOSPHATE AMINOTRANSFERASE COMPLEXED WITH HISTIDINOL-PHOSPHATE | | Descriptor: | HISTIDINOL-PHOSPHATE AMINOTRANSFERASE, PHOSPHORIC ACID MONO-[2-AMINO-3-(3H-IMIDAZOL-4-YL)-PROPYL]ESTER, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Haruyama, K, Nakai, T, Miyahara, I, Hirotsu, K, Mizuguchi, H, Hayashi, H, Kagamiyama, H. | | Deposit date: | 2000-11-30 | | Release date: | 2001-04-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme.
Biochemistry, 40, 2001
|
|
1GDE
 
 | | CRYSTAL STRUCTURE OF PYROCOCCUS PROTEIN A-1 E-FORM | | Descriptor: | ASPARTATE AMINOTRANSFERASE, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Ura, H, Harata, K, Matsui, I, Kuramitsu, S. | | Deposit date: | 2000-09-23 | | Release date: | 2001-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature dependence of the enzyme-substrate recognition mechanism.
J.Biochem., 129, 2001
|
|
5QQQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of human ALAS2A in complex with Z2856434857 | | Descriptor: | 3-[(thiomorpholin-4-yl)methyl]phenol, 5-aminolevulinate synthase, erythroid-specific, ... | | Authors: | Bezerra, G.A, Foster, W, Bailey, H, Shrestha, L, Krojer, T, Talon, R, Brandao-Neto, J, Douangamath, A, Nicola, B.B, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Brennan, P.E, Yue, W.W. | | Deposit date: | 2019-05-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
