3G9A
 
 | | Green fluorescent protein bound to minimizer nanobody | | Descriptor: | Green fluorescent protein, Minimizer | | Authors: | Kirchhofer, A, Helma, J, Schmidthals, K, Frauer, C, Cui, S, Karcher, A, Pellis, M, Muyldermans, S, Delucci, C.C, Cardoso, M.C, Leonhardt, H, Hopfner, K.-P, Rothbauer, U. | | Deposit date: | 2009-02-13 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.614 Å) | | Cite: | Modulation of protein properties in living cells using nanobodies
Nat.Struct.Mol.Biol., 17, 2010
|
|
6FPB
 
 | | Crystal structure of anti-mTFP1 DARPin 1238_G01 in space group I4 | | Descriptor: | CHLORIDE ION, DARPin 1238_G01 | | Authors: | Jakob, R.P, Vigano, M.A, Bieli, D, Matsuda, S, Schaefer, J.V, Pluckthun, A, Affolter, M, Maier, T. | | Deposit date: | 2018-02-09 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | DARPins recognizing mTFP1 as novel reagents forin vitroandin vivoprotein manipulations.
Biol Open, 7, 2018
|
|
6PFU
 
 | |
8I4J
 
 | |
6A1U
 
 | | Charcot-Leyden crystal protein/Galectin-10 variant E33D | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
4EN1
 
 | |
6DEJ
 
 | |
3SSP
 
 | |
6A1S
 
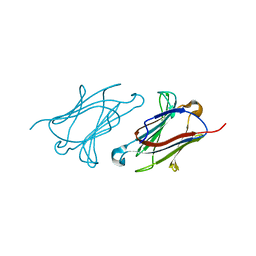 | | Charcot-Leyden crystal protein/Galectin-10 variant E33A | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
6A63
 
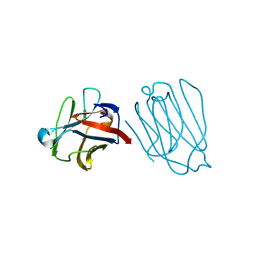 | | Placental protein 13/galectin-13 variant R53HH57R with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A64
 
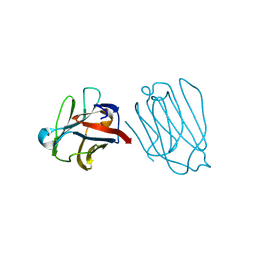 | | Placental protein 13/galectin-13 variant R53HR55NH57RD33G with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
8A6G
 
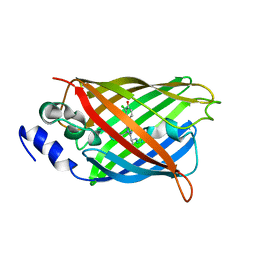 | |
8A6R
 
 | |
8A6O
 
 | |
8A6S
 
 | |
8A6N
 
 | |
8A6P
 
 | |
8A6Q
 
 | |
6A1Y
 
 | | Charcot-Leyden crystal protein/Galectin-10 variant Y35A | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
7UGS
 
 | |
7AMF
 
 | |
7AMB
 
 | |
3IR8
 
 | | Red fluorescent protein mKeima at pH 7.0 | | Descriptor: | Large stokes shift fluorescent protein | | Authors: | Henderson, J.N, Osborn, M.F, Koon, N, Gepshtein, R, Huppert, D, Remington, S.J. | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Excited state proton transfer in the red fluorescent protein mKeima.
J.Am.Chem.Soc., 131, 2009
|
|
2YE1
 
 | | X-ray structure of the cyan fluorescent proteinmTurquoise-GL (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN, MAGNESIUM ION | | Authors: | von Stetten, D, Noirclerc-Savoye, M, Goedhart, J, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural Characterization of the Cyan Fluorescent Protein Mturquoise-Gl
To be Published
|
|
4JZK
 
 | |
