2D6B
 
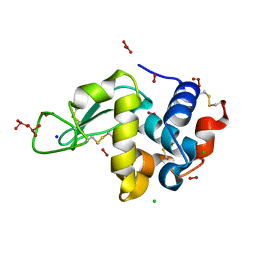 | | Novel Bromate Species trapped within a Protein Crystal | | Descriptor: | BROMIC ACID, CHLORIDE ION, SODIUM ION, ... | | Authors: | Ondracek, J, Mesters, J.R. | | Deposit date: | 2005-11-10 | | Release date: | 2005-11-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | An ensemble of crystallographic models enables the description of novel bromate-oxoanion species trapped within a protein crystal
Acta Crystallogr.,Sect.D, 62, 2006
|
|
2D6C
 
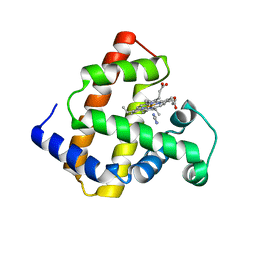 | | Crystal structure of myoglobin reconstituted with iron porphycene | | Descriptor: | IMIDAZOLE, Myoglobin, PORPHYCENE CONTAINING FE | | Authors: | Hayashi, T, Murata, D, Makino, M, Sugimoto, H, Matsuo, T, Sato, H, Shiro, Y, Hisaeda, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-11 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure and peroxidase activity of myoglobin reconstituted with iron porphycene
Inorg.Chem., 45, 2006
|
|
2D6F
 
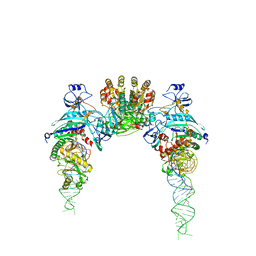 | |
2D6K
 
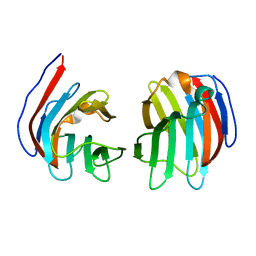 | | Crystal structure of mouse galectin-9 N-terminal CRD (crystal form 1) | | Descriptor: | lectin, galactose binding, soluble 9 | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6L
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD (crystal form 2) | | Descriptor: | lectin, galactose binding, soluble 9 | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6M
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, lectin, galactose binding, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6N
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with N-acetyllactosamine | | Descriptor: | beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin, galactose binding, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6O
 
 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with N-acetyllactosamine dimer | | Descriptor: | GLYCEROL, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, lectin, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6P
 
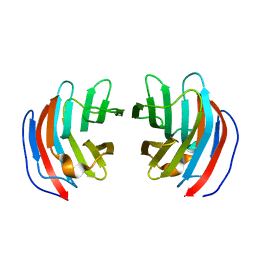 | | Crystal structure of mouse galectin-9 N-terminal CRD in complex with T-antigen | | Descriptor: | beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, lectin, galactose binding, ... | | Authors: | Nagae, M, Nishi, N, Nakamura, T, Murata, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2005-11-14 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Galectin-9 N-terminal Carbohydrate Recognition Domain from Mus musculus Reveals the Basic Mechanism of Carbohydrate Recognition
J.Biol.Chem., 281, 2006
|
|
2D6Y
 
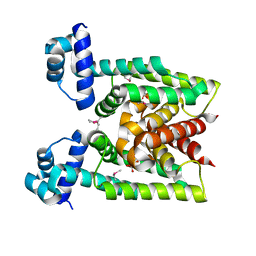 | | Crystal Structure of transcriptional factor SCO4008 from Streptomyces coelicolor A3(2) | | Descriptor: | L(+)-TARTARIC ACID, putative tetR family regulatory protein | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2005-11-15 | | Release date: | 2006-10-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SCO4008, a Putative TetR Transcriptional Repressor from Streptomyces coelicolor A3(2), Regulates Transcription of sco4007 by Multidrug Recognition.
J.Mol.Biol., 425, 2013
|
|
2D73
 
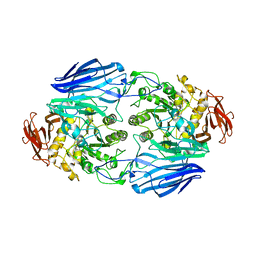 | | Crystal Structure Analysis of SusB | | Descriptor: | CALCIUM ION, alpha-glucosidase SusB | | Authors: | Kitamura, M, Yao, M. | | Deposit date: | 2005-11-15 | | Release date: | 2007-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron.
J.Biol.Chem., 283, 2008
|
|
2D74
 
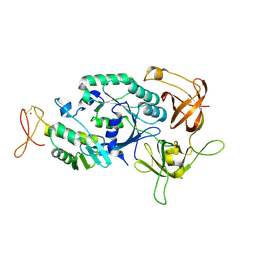 | | Crystal structure of translation initiation factor aIF2betagamma heterodimer | | Descriptor: | Translation initiation factor 2 beta subunit, Translation initiation factor 2 gamma subunit, ZINC ION | | Authors: | Sokabe, M, Yao, M, Sakai, N, Toya, S, Tanaka, I. | | Deposit date: | 2005-11-16 | | Release date: | 2006-07-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of archaeal translational initiation factor 2 betagamma-GDP reveals significant conformational change of the beta-subunit and switch 1 region.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2D7C
 
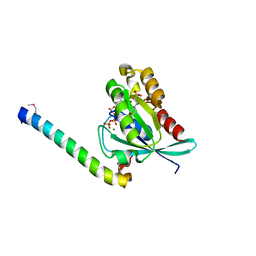 | | Crystal structure of human Rab11 in complex with FIP3 Rab-binding domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shiba, T, Koga, H, Shin, H.W, Kawasaki, M, Kato, R, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2005-11-16 | | Release date: | 2006-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for Rab11-dependent membrane recruitment of a family of Rab11-interacting protein 3 (FIP3)/Arfophilin-1.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2D7D
 
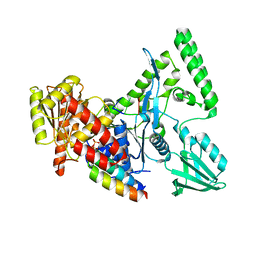 | |
2D7E
 
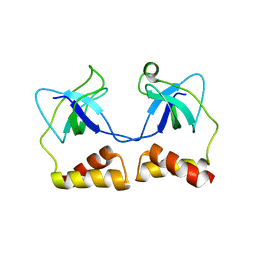 | | Crystal structure of N-terminal domain of PriA from E.coli | | Descriptor: | Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-18 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7F
 
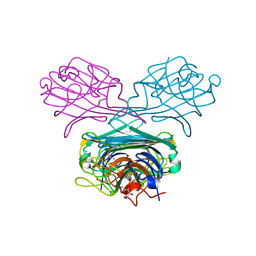 | | Crystal structure of A lectin from canavalia gladiata seeds complexed with alpha-methyl-mannoside and alpha-aminobutyric acid | | Descriptor: | CALCIUM ION, Concanavalin A, D-ALPHA-AMINOBUTYRIC ACID, ... | | Authors: | Delatorre, P, Rocha, B.A.M, Souza, E.P, Freitas, B.T, Moreno, F.B.B.M, Sampaio, A.H, Azevedo Jr, W.F, Cavada, B.S. | | Deposit date: | 2005-11-19 | | Release date: | 2006-12-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of a lectin from Canavalia gladiata seeds: new structural insights for old molecules
Bmc Struct.Biol., 7, 2007
|
|
2D7G
 
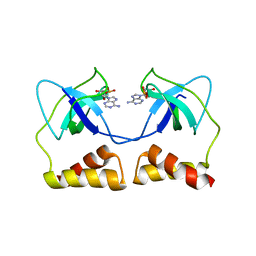 | | Crystal structure of the aa complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*AP*A)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7H
 
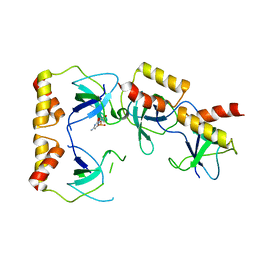 | | Crystal structure of the ccc complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*CP*CP*C)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7I
 
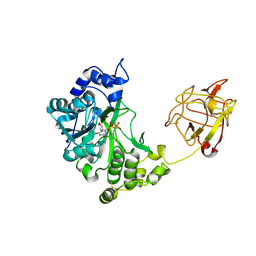 | | Crystal structure of pp-GalNAc-T10 with UDP, GalNAc and Mn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kubota, T, Shiba, T, Sugioka, S, Kato, R, Wakatsuki, S, Narimatsu, H. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10)
J.Mol.Biol., 359, 2006
|
|
2D7J
 
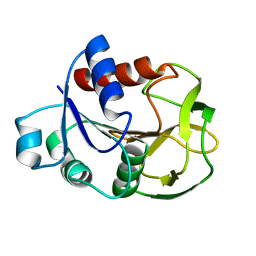 | | Crystal Structure Analysis of Glutamine Amidotransferase from Pyrococcus horikoshii OT3 | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit A | | Authors: | Maruoka, S, Lee, W.C, Kamo, M, Kudo, N, Nagata, K, Tanokura, M. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal structure of glutamine amidotransferase from Pyrococcus horikoshii OT3
PROC.JPN.ACAD.,SER.B, 81, 2005
|
|
2D7L
 
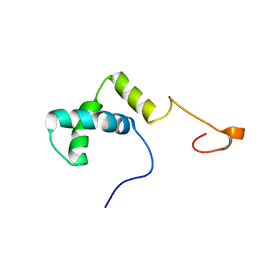 | | Solution structure of the HMG box domain from human WD repeat and HMG-box DNA binding protein 1 | | Descriptor: | WD repeat and HMG-box DNA binding protein 1 | | Authors: | Tomizawa, T, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Kamatari, Y.O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HMG box domain from human WD repeat and HMG-box DNA binding protein 1
To be Published
|
|
2D7M
 
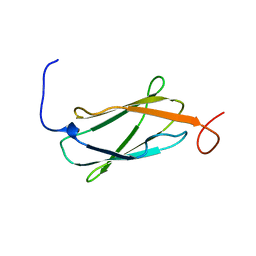 | | Solution structure of the 14th Filamin domain from human Filamin C | | Descriptor: | Filamin-C | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 14th Filamin domain from human Filamin C
To be Published
|
|
2D7N
 
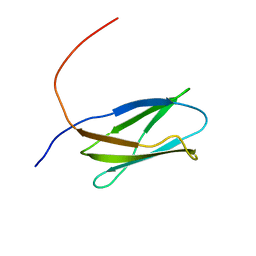 | | Solution structure of the 16th Filamin domain from human Filamin C | | Descriptor: | Filamin-C | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 16th Filamin domain from human Filamin C
To be Published
|
|
2D7O
 
 | | Solution structure of the 17th Filamin domain from human Filamin C | | Descriptor: | Filamin-C | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 17th Filamin domain from human Filamin C
To be Published
|
|
2D7P
 
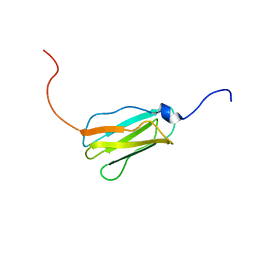 | | Solution structure of the 22th Filamin domain from human Filamin C | | Descriptor: | Filamin-C | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 22th Filamin domain from human Filamin C
To be Published
|
|
