6HE8
 
 | | PAN-proteasome in state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Majumder, P, Rudack, T, Beck, F, Baumeister, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.86 Å) | | Cite: | Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6J2C
 
 | | Yeast proteasome in translocation competent state (C3-a) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Cong, Y. | | Deposit date: | 2019-01-01 | | Release date: | 2019-03-13 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
6J2X
 
 | | Yeast proteasome in resting state (C1-a) | | Descriptor: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASOME REGULATORY SUBUNIT RPN5, 26S proteasome complex subunit SEM1, ... | | Authors: | Cong, Y. | | Deposit date: | 2019-01-03 | | Release date: | 2019-03-13 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
6JPQ
 
 | | CryoEM structure of Abo1 hexamer - ADP complex | | Descriptor: | Uncharacterized AAA domain-containing protein C31G5.19 | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.44 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
6JPU
 
 | | CryoEM structure of Abo1 hexamer - apo complex | | Descriptor: | Uncharacterized AAA domain-containing protein C31G5.19 | | Authors: | Cho, C, Jang, J, Song, J.J. | | Deposit date: | 2019-03-28 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural basis of nucleosome assembly by the Abo1 AAA+ ATPase histone chaperone.
Nat Commun, 10, 2019
|
|
6J2Q
 
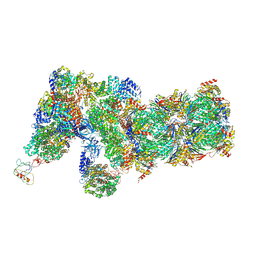 | | Yeast proteasome in Ub-accepted state (C1-b) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Cong, Y. | | Deposit date: | 2019-01-02 | | Release date: | 2019-03-13 | | Last modified: | 2019-04-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
8FS5
 
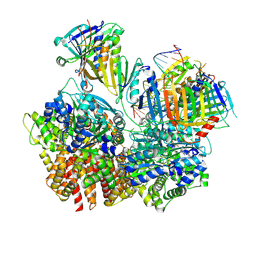 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 3 (open 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS4
 
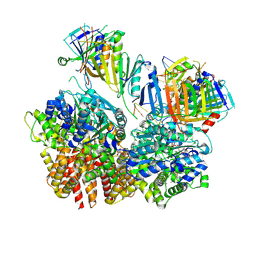 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 2 (open 9-1-1 ring and flexibly bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS3
 
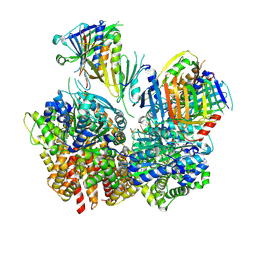 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 1 (open 9-1-1 and shoulder bound DNA only) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS8
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 5-nt gapped DNA (9-1-1 encircling fully bound DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS7
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 5 (closed 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FS6
 
 | | Structure of S. cerevisiae Rad24-RFC loading the 9-1-1 clamp onto a 10-nt gapped DNA in step 4 (partially closed 9-1-1 and stably bound chamber DNA) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, DDC1 isoform 1, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2023-01-09 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of 9-1-1 DNA checkpoint clamp loading at gaps from start to finish and ramification to biology.
Biorxiv, 2023
|
|
8FCN
 
 | | Cryo-EM structure of p97:UBXD1 VIM-only state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8G28
 
 | | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae. | | Descriptor: | ATPase, AAA family, CHLORIDE ION | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-02-03 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of the C-terminal Fragment of AAA ATPase from Streptococcus pneumoniae.
To Be Published
|
|
8DR4
 
 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) without NTD | | Descriptor: | DNA (5'-D(P*AP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR7
 
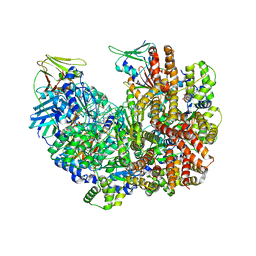 | | Open state of RFC:PCNA bound to a nicked dsDNA | | Descriptor: | DNA (26-MER), DNA (5'-D(P*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR3
 
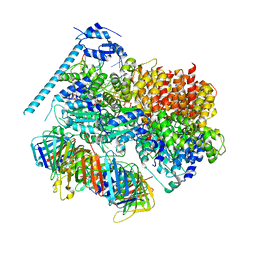 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR5
 
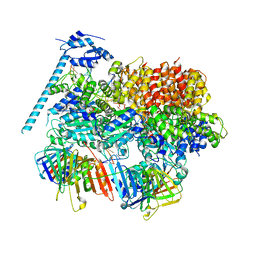 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD | | Descriptor: | DNA (5'-D(P*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR1
 
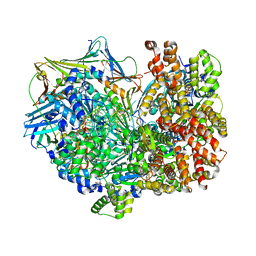 | | Consensus closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DQW
 
 | |
8GLV
 
 | |
8FCT
 
 | | Cryo-EM structure of p97:UBXD1 lariat mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCP
 
 | | Cryo-EM structure of p97:UBXD1 para state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCL
 
 | | Cryo-EM structure of p97:UBXD1 closed state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCR
 
 | | Cryo-EM structure of p97:UBXD1 H4-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | Authors: | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
