3S83
 
 | | EAL domain of phosphodiesterase PdeA | | Descriptor: | GGDEF family protein, POTASSIUM ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Massa, C, Schirmer, T, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of EAL domain from Caulobacter crescentus CB15
To be Published
|
|
3SC6
 
 | | 2.65 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose reductase (rfbD) from Bacillus anthracis str. Ames in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, dTDP-4-dehydrorhamnose reductase | | Authors: | Halavaty, A.S, Kuhn, M, Shuvalova, L, Minasov, G, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-07 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme dTDP-4-dehydrorhamnose reductase (RfbD).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3SD7
 
 | | 1.7 Angstrom Resolution Crystal Structure of Putative Phosphatase from Clostridium difficile | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative phosphatase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Resolution Crystal Structure of Putative Phosphatase from Clostridium difficile.
TO BE PUBLISHED
|
|
9GB9
 
 | |
7FK3
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04A09 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8, methyl N-(benzenesulfonyl)-N-methylglycinate | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
3SEF
 
 | | 2.4 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate and NADPH | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Shikimate 5-dehydrogenase | | Authors: | Halavaty, A.S, Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-10 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom resolution crystal structure of shikimate 5-dehydrogenase (aroE) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with shikimate and NADPH
To be Published
|
|
7FJV
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03H10 from the F2X-Universal Library | | Descriptor: | (3S)-3-amino-4-(2-fluorophenyl)butanoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FJW
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03H12 from the F2X-Universal Library | | Descriptor: | 6-[(pyridin-2-yl)sulfanyl]pyridin-3-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FK1
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04A06 from the F2X-Universal Library | | Descriptor: | (3R)-3-(4-chlorobenzene-1-sulfonyl)butanoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FK0
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04A05 from the F2X-Universal Library | | Descriptor: | 2-amino-N-(3-fluoro-4-methylphenyl)ethane-1-sulfonamide, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FJU
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P03H10 from the F2X-Universal Library | | Descriptor: | (3S)-3-amino-4-(2-fluorophenyl)butanoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FJY
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04A02 from the F2X-Universal Library | | Descriptor: | 1-[(3-aminophenyl)methyl]piperidin-4-ol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FJZ
 
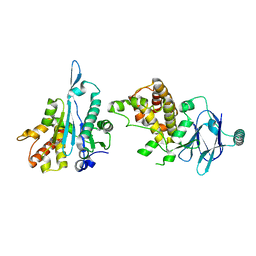 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04A04 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, D-phenylalaninamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FJX
 
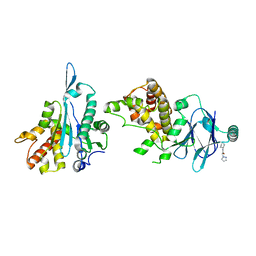 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04A01 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N-cyclohexyl-2-(1H-pyrazol-1-yl)acetamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FK2
 
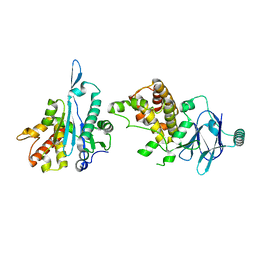 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04A07 from the F2X-Universal Library | | Descriptor: | 4-(thiophene-2-sulfonyl)morpholine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7NM6
 
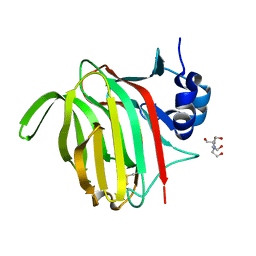 | | Crystal structure of Paradendryphiella salina PL7A alginate lyase mutant Y223F in complex with di-mannuronic acid | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alginate lyase (PL7), beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Fredslund, F, Welner, D.H, Wilkens, C. | | Deposit date: | 2021-02-23 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Unraveling the molecular mechanism of polysaccharide lyases for efficient alginate degradation.
Nat Commun, 16, 2025
|
|
7NL3
 
 | |
7NPP
 
 | | Crystal structure of Paradendryphiella salina PL7A alginate lyase mutant Y223F in complex with penta-mannuronic acid | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alginate lyase (PL7), beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Fredslund, F, Welner, D.W, Wilkens, C. | | Deposit date: | 2021-02-28 | | Release date: | 2022-03-23 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Unraveling the molecular mechanism of polysaccharide lyases for efficient alginate degradation.
Nat Commun, 16, 2025
|
|
7NY3
 
 | | Crystal structure of Paradendryphiella salina PL7A alginate lyase mutant Y223F in complex with tetra-mannuronic acid | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alginate lyase (PL7), beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Fredslund, F, Welner, D.W, Wilkens, C. | | Deposit date: | 2021-03-20 | | Release date: | 2022-03-30 | | Last modified: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Unraveling the molecular mechanism of polysaccharide lyases for efficient alginate degradation.
Nat Commun, 16, 2025
|
|
7FKC
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04C10 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N,N~2~-dimethyl-N-(pyridin-2-yl)glycinamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FLT
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P05G04 from the F2X-Universal Library | | Descriptor: | (1S)-1-(3-fluoro-4-methoxyphenyl)ethan-1-amine, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FNT
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07E12 from the F2X-Universal Library | | Descriptor: | 2-{[(3-bromophenyl)methyl]amino}ethan-1-ol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FNQ
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P07E01 from the F2X-Universal Library | | Descriptor: | (3S)-3-methyl-5-oxo-5-[(1,3-thiazol-2-yl)amino]pentanoic acid, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FOU
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08E01 from the F2X-Universal Library | | Descriptor: | (1R)-1-(4-fluorophenyl)-2-[(propan-2-yl)amino]ethan-1-ol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7FP3
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P08F11 from the F2X-Universal Library | | Descriptor: | A1 cistron-splicing factor AAR2, N,N~2~-diethyl-N~2~-(2-methylpyrimidin-4-yl)glycinamide, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
