1CCB
 
 | | THE ASP-HIS-FE TRIAD OF CYTOCHROME C PEROXIDASE CONTROLS THE REDUCTION POTENTIAL, ELECTRONIC STRUCTURE, AND COUPLING OF THE TRYPTOPHAN FREE-RADICAL TO THE HEME | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Goodin, D.B, Mcree, D.E. | | Deposit date: | 1993-01-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Asp-His-Fe triad of cytochrome c peroxidase controls the reduction potential, electronic structure, and coupling of the tryptophan free radical to the heme.
Biochemistry, 32, 1993
|
|
1CCC
 
 | | THE ASP-HIS-FE TRIAD OF CYTOCHROME C PEROXIDASE CONTROLS THE REDUCTION POTENTIAL, ELECTRONIC STRUCTURE, AND COUPLING OF THE TRYPTOPHAN FREE-RADICAL TO THE HEME | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Goodin, D.B, Mcree, D.E. | | Deposit date: | 1993-01-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Asp-His-Fe triad of cytochrome c peroxidase controls the reduction potential, electronic structure, and coupling of the tryptophan free radical to the heme.
Biochemistry, 32, 1993
|
|
1CCD
 
 | | REFINED STRUCTURE OF RAT CLARA CELL 17 KDA PROTEIN AT 3.0 ANGSTROMS RESOLUTION | | Descriptor: | CLARA CELL 17 kD PROTEIN, SULFATE ION | | Authors: | Umland, T.C, Swaminathan, S, Furey, W, Singh, G, Pletcher, J, Sax, M. | | Deposit date: | 1991-09-17 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Refined structure of rat Clara cell 17 kDa protein at 3.0 A resolution.
J.Mol.Biol., 224, 1992
|
|
1CCE
 
 | | CONSTRUCTION OF A BIS-AQUO HEME ENZYME AND REPLACEMENT WITH EXOGENOUS LIGAND | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mcree, D.E, Jensen, G.M, Fitzgerald, M.M, Siegel, H.A, Goodin, D.B. | | Deposit date: | 1994-05-04 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Construction of a bisaquo heme enzyme and binding by exogenous ligands.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1CCF
 
 | | How an Epidermal Growth Factor (EGF)-Like Domain Binds Calcium-High Resolution NMR Structure of the Calcium Form of the NH2-Terminal EGF-Like Domain in Coagulation Factor X | | Descriptor: | COAGULATION FACTOR X | | Authors: | Selander-Sunnerhagen, M, Ullner, M, Persson, M, Teleman, O, Stenflo, J, Drakenberg, T. | | Deposit date: | 1993-05-19 | | Release date: | 1994-05-31 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | How an epidermal growth factor (EGF)-like domain binds calcium. High resolution NMR structure of the calcium form of the NH2-terminal EGF-like domain in coagulation factor X.
J.Biol.Chem., 267, 1992
|
|
1CCG
 
 | | CONSTRUCTION OF A BIS-AQUO HEME ENZYME AND REPLACEMENT WITH EXOGENOUS LIGAND | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mcree, D.E, Jensen, G.M, Fitzgerald, M.M, Siegel, H.A, Goodin, D.B. | | Deposit date: | 1994-05-04 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Construction of a bisaquo heme enzyme and binding by exogenous ligands.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1CCH
 
 | |
1CCI
 
 | | HOW FLEXIBLE ARE PROTEINS? TRAPPING OF A FLEXIBLE LOOP | | Descriptor: | 2,3-DIMETHYLIMIDAZOLIUM ION, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cao, Y, Musah, R.A, Wilcox, S.K, Goodin, D.B, Mcree, D.E. | | Deposit date: | 1996-12-18 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A ligand-gated, hinged loop rearrangement opens a channel to a buried artificial protein cavity.
Nat.Struct.Biol., 3, 1996
|
|
1CCJ
 
 | | CONFORMER SELECTION BY LIGAND BINDING OBSERVED WITH PROTEIN CRYSTALLOGRAPHY | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cao, Y, Musah, R.A, Wilcox, S.K, Goodin, D.B, Mcree, D.E. | | Deposit date: | 1997-01-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein conformer selection by ligand binding observed with crystallography.
Protein Sci., 7, 1998
|
|
1CCK
 
 | | ALTERING SUBSTRATE SPECIFICITY OF CYTOCHROME C PEROXIDASE TOWARDS A SMALL MOLECULAR SUBSTRATE PEROXIDASE BY SUBSTITUTING TYROSINE FOR PHE 202 | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cao, Y, Musah, R.A, Wilcox, S.K, Goodin, D.B, Mcree, D.E. | | Deposit date: | 1998-01-02 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of recombinant pea cytosolic ascorbate peroxidase.
Biochemistry, 34, 1995
|
|
1CCL
 
 | |
1CCM
 
 | | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | | Descriptor: | CRAMBIN | | Authors: | Bonvin, A.M.J.J, Rullmann, J.A.C, Lamerichs, R.M.J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-04-14 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | "Ensemble" iterative relaxation matrix approach: a new NMR refinement protocol applied to the solution structure of crambin.
Proteins, 15, 1993
|
|
1CCN
 
 | | DIRECT NOE REFINEMENT OF CRAMBIN FROM 2D NMR DATA USING A SLOW-COOLING ANNEALING PROTOCOL | | Descriptor: | CRAMBIN | | Authors: | Bonvin, A.M.J.J, Rullmann, J.A.C, Lamerichs, R.M.J.N, Boelens, R, Kaptein, R. | | Deposit date: | 1993-04-14 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Direct NOE refinement of biomolecular structures using 2D NMR data
J.Biomol.NMR, 1, 1991
|
|
1CCP
 
 | | X-RAY STRUCTURES OF RECOMBINANT YEAST CYTOCHROME C PEROXIDASE AND THREE HEME-CLEFT MUTANTS PREPARED BY SITE-DIRECTED MUTAGENESIS | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, YEAST CYTOCHROME C PEROXIDASE | | Authors: | Wang, J, Mauro, J.M, Edwards, S.L, Oatley, S.J, Fishel, L.A, Ashford, V.A, Xuong, N.-H, Kraut, J. | | Deposit date: | 1990-02-28 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of recombinant yeast cytochrome c peroxidase and three heme-cleft mutants prepared by site-directed mutagenesis.
Biochemistry, 29, 1990
|
|
1CCQ
 
 | |
1CCR
 
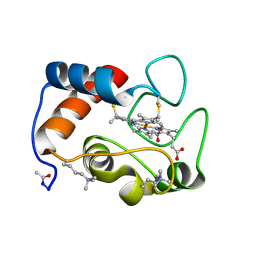 | | STRUCTURE OF RICE FERRICYTOCHROME C AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Ochi, H, Hata, Y, Tanaka, N, Kakudo, M, Sakurai, T, Aihara, S, Morita, Y. | | Deposit date: | 1983-03-14 | | Release date: | 1983-04-21 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of rice ferricytochrome c at 2.0 A resolution.
J.Mol.Biol., 166, 1983
|
|
1CCS
 
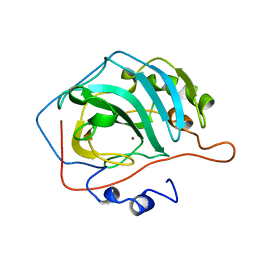 | |
1CCT
 
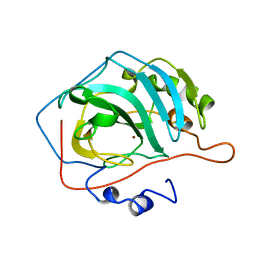 | |
1CCU
 
 | |
1CCV
 
 | |
1CCW
 
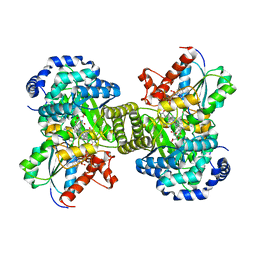 | |
1CCZ
 
 | | CRYSTAL STRUCTURE OF THE CD2-BINDING DOMAIN OF CD58 (LYMPHOCYTE FUNCTION-ASSOCIATED ANTIGEN 3) AT 1.8-A RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CD58) | | Authors: | Ikemizu, S, Sparks, L.M, Van Der Merwe, P.A, Harlos, K, Stuart, D.I, Jones, E.Y, Davis, S.J. | | Deposit date: | 1999-03-02 | | Release date: | 1999-04-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the CD2-binding domain of CD58 (lymphocyte function-associated antigen 3) at 1.8-A resolution.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CD0
 
 | | STRUCTURE OF HUMAN LAMDA-6 LIGHT CHAIN DIMER JTO | | Descriptor: | PROTEIN (JTO, A VARIABLE DOMAIN FROM LAMBDA-6 TYPE IMMUNOGLOBULIN LIGHT CHAIN) | | Authors: | Pokkuluri, P.R, Solomon, A, Weiss, D.T, Stevens, F.J, Schiffer, M. | | Deposit date: | 1999-03-05 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tertiary structure of human lambda 6 light chains.
Amyloid, 6, 1999
|
|
1CD1
 
 | |
1CD2
 
 | | LIGAND INDUCED CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURES OF PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE COMPLEXES WITH FOLATE AND NADP+ | | Descriptor: | DIHYDROFOLATE REDUCTASE, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Blakley, R.L, Gangjee, A. | | Deposit date: | 1999-03-05 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced conformational changes in the crystal structures of Pneumocystis carinii dihydrofolate reductase complexes with folate and NADP+.
Biochemistry, 38, 1999
|
|
