7OT4
 
 | |
7OU3
 
 | | Crystal structure of Tga-AGOG, an 8-oxoguanine DNA glycosylase from Thermococcus gammatolerans | | Descriptor: | CHLORIDE ION, GLYCEROL, N-glycosylase/DNA lyase | | Authors: | Coste, F, Confalonieri, F, Castaing, B. | | Deposit date: | 2021-06-11 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and functional determinants of the archaeal 8-oxoguanine-DNA glycosylase AGOG for DNA damage recognition and processing.
Nucleic Acids Res., 50, 2022
|
|
3H54
 
 | | Crystal Structure of human alpha-N-acetylgalactosaminidase,complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Clark, N.E, Garman, S.C. | | Deposit date: | 2009-04-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 1.9 a structure of human alpha-N-acetylgalactosaminidase: The molecular basis of Schindler and Kanzaki diseases
J.Mol.Biol., 393, 2009
|
|
4K86
 
 | |
6D4C
 
 | | Crystal structure of Candida boidinii formate dehydrogenase V123G mutant complexed with NAD+ and azide | | Descriptor: | AZIDE ION, CHLORIDE ION, Formate dehydrogenase, ... | | Authors: | Guo, Q, Ye, H, Gakhar, L, Cheatum, C.M, Kohen, A. | | Deposit date: | 2018-04-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Oscillatory Active-site Motions Correlate with Kinetic Isotope Effects in Formate Dehydrogenase
Acs Catalysis, 2019
|
|
4K40
 
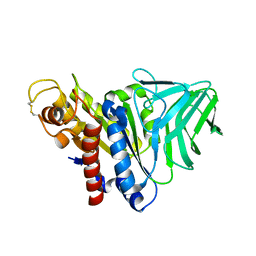 | | Peptidoglycan O-acetylesterase in action, 0 min | | Descriptor: | GDSL-like Lipase/Acylhydrolase family protein | | Authors: | Williams, A.H, Gompert Boneca, I. | | Deposit date: | 2013-04-11 | | Release date: | 2014-09-03 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Visualization of a substrate-induced productive conformation of the catalytic triad of the Neisseria meningitidis peptidoglycan O-acetylesterase reveals mechanistic conservation in SGNH esterase family members.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3D9G
 
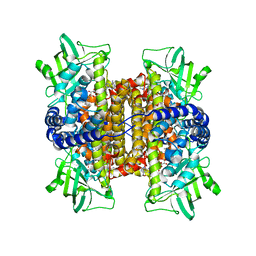 | | Nitroalkane oxidase: wild type crystallized in a trapped state forming a cyanoadduct with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Nitroalkane oxidase, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
3DES
 
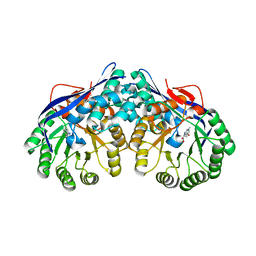 | | Crystal structure of dipeptide epimerase from Thermotoga maritima complexed with L-Ala-L-Phe dipeptide | | Descriptor: | ALANINE, MAGNESIUM ION, Muconate cycloisomerase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a dipeptide epimerase enzymatic function guided by homology modeling and virtual screening.
Structure, 16, 2008
|
|
3DAN
 
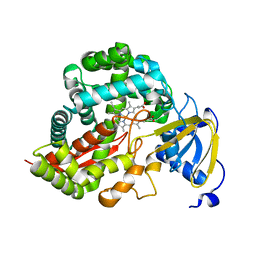 | | Crystal Structure of Allene oxide synthase | | Descriptor: | Cytochrome P450 74A2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, L, Wang, X. | | Deposit date: | 2008-05-29 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5XB8
 
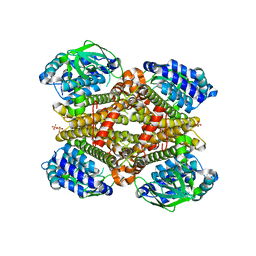 | | Crystal structure of dibenzothiophene monooxygenase (TdsC) from Paenibacillus sp. A11-2 | | Descriptor: | SULFATE ION, Thermophilic dibenzothiophene desulfurization enzyme C | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
3DBM
 
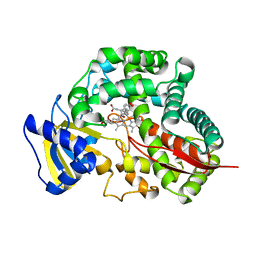 | | Crystal Structure of Allene oxide synthase | | Descriptor: | (9E,11E,13S)-13-hydroxyoctadeca-9,11-dienoic acid, Cytochrome P450 74A2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, L, Wang, X. | | Deposit date: | 2008-06-01 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3GL0
 
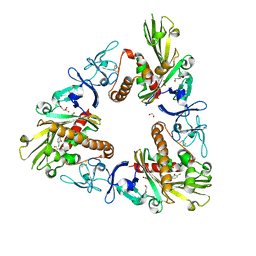 | | Crystal structure of dicamba monooxygenase bound to 3,6 dichlorosalicylic acid (DCSA) | | Descriptor: | 1,2-ETHANEDIOL, 3,6-dichloro-2-hydroxybenzoic acid, DdmC, ... | | Authors: | Wilson, M.A, Dumitru, R, Jiang, W.Z, Weeks, D.P. | | Deposit date: | 2009-03-11 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dicamba monooxygenase: a Rieske nonheme oxygenase that catalyzes oxidative demethylation.
J.Mol.Biol., 392, 2009
|
|
2Q9Z
 
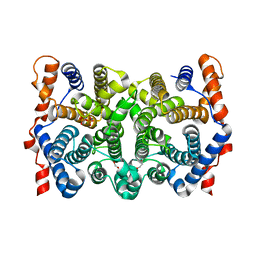 | | Trichodiene synthase: Complex with inorganic pyrophosphate resulting from the reaction with 2-fluorofarnesyl diphosphate | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Zhao, Y, Coates, R.M, Koyama, T, Cane, D.E, Christianson, D.W. | | Deposit date: | 2007-06-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Exploring biosynthetic diversity with trichodiene synthase.
Arch.Biochem.Biophys., 466, 2007
|
|
6HWZ
 
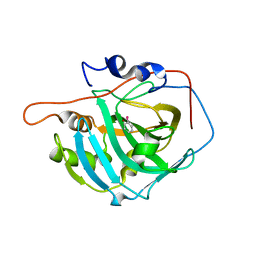 | |
2QVP
 
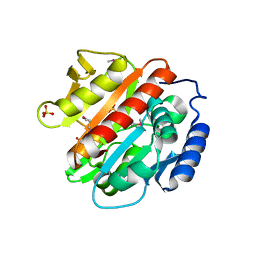 | |
5XDD
 
 | | Crystal structure of tertiary complex of TdsC from Paenibacillus sp. A11-2 with FMN and Indole | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, INDOLE, ... | | Authors: | Hino, T, Hamamoto, H, Ohshiro, T, Nagano, S. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of TdsC, a dibenzothiophene monooxygenase from the thermophile Paenibacillus sp. A11-2, reveal potential for expanding its substrate selectivity.
J. Biol. Chem., 292, 2017
|
|
6D5D
 
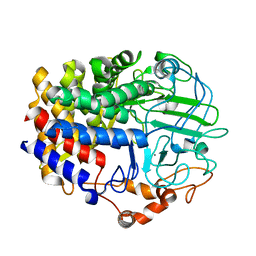 | |
3GOB
 
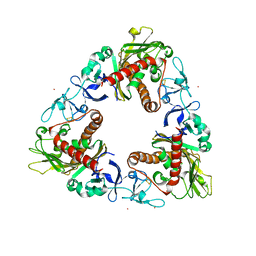 | | Crystal Structure of Dicamba Monooxygenase with Non-heme Cobalt and DCSA | | Descriptor: | 3,6-dichloro-2-hydroxybenzoic acid, COBALT (II) ION, DdmC, ... | | Authors: | Rydel, T.J, Sturman, E.J, Moshiri, F, Brown, G.R, Qi, Y. | | Deposit date: | 2009-03-18 | | Release date: | 2009-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Dicamba monooxygenase: structural insights into a dynamic Rieske oxygenase that catalyzes an exocyclic monooxygenation.
J.Mol.Biol., 392, 2009
|
|
2G82
 
 | |
4ONQ
 
 | |
3GP5
 
 | |
3DBL
 
 | |
6DBG
 
 | |
3DDF
 
 | | GOLGI MANNOSIDASE II complex with (3R,4R,5R)-3,4-Dihydroxy-5-({[(1R)-2-hydroxy-1 phenylethyl]amino}methyl) pyrrolidin-2-one | | Descriptor: | (3R,4R,5R)-3,4-dihydroxy-5-({[(1R)-2-hydroxy-1-phenylethyl]amino}methyl)pyrrolidin-2-one, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Kuntz, D.A, Rose, D.R, Hoffman, D. | | Deposit date: | 2008-06-05 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Functionalized pyrrolidine inhibitors of human type II alpha-mannosidases as anti-cancer agents: optimizing the fit to the active site
Bioorg.Med.Chem., 16, 2008
|
|
3DHG
 
 | | Crystal Structure of Toluene 4-Monoxygenase Hydroxylase | | Descriptor: | AZIDE ION, CALCIUM ION, FE (III) ION, ... | | Authors: | Bailey, L.J, Mccoy, J.G, Phillips Jr, G.N, Fox, B.G. | | Deposit date: | 2008-06-17 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural consequences of effector protein complex formation in a diiron hydroxylase.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
