2AN5
 
 | | Structure of human PNMT complexed with S-adenosyl-homocysteine and an inhibitor, trans-(1S,2S)-2-amino-1-tetralol | | Descriptor: | PHOSPHATE ION, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gee, C.L, Tyndall, J.D.A, Grunewald, G.L, Wu, Q, McLeish, M.J, Martin, J.L. | | Deposit date: | 2005-08-11 | | Release date: | 2006-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mode of binding of methyl acceptor substrates to the adrenaline-synthesizing enzyme phenylethanolamine N-methyltransferase: implications for catalysis
Biochemistry, 44, 2005
|
|
4IEY
 
 | | Cys-persulfenate bound Cysteine Dioxygenase at pH 7.0 in the presence of Cys, home-source structure | | Descriptor: | Cysteine dioxygenase type 1, FE (II) ION, S-HYDROPEROXYCYSTEINE | | Authors: | Driggers, C.M, Cooley, R.B, Karplus, P.A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Cysteine Dioxygenase Structures from pH4 to 9: Consistent Cys-Persulfenate Formation at Intermediate pH and a Cys-Bound Enzyme at Higher pH.
J.Mol.Biol., 425, 2013
|
|
6MWK
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (IspF) Burkholderia pseudomallei in complex with ligand HGN-0883 | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 4-amino-N-(6-methoxypyrimidin-4-yl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of Burkholderia pseudomallei IspF in complex with sulfapyridine, sulfamonomethoxine, ethoxzolamide and acetazolamide
Acta Crystallogr.,Sect.F, 81, 2025
|
|
3QI6
 
 | |
1KVX
 
 | |
6MWI
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (IspF) Burkholderia pseudomallei in complex with ligand HGN-0456 | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 6-ethoxy-1,3-benzothiazole-2-sulfonamide, ACETATE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of Burkholderia pseudomallei IspF in complex with sulfapyridine, sulfamonomethoxine, ethoxzolamide and acetazolamide
Acta Crystallogr.,Sect.F, 81, 2025
|
|
3WJY
 
 | |
4QOC
 
 | |
3OKN
 
 | | Crystal structure of S25-39 in complex with Kdo(2.4)Kdo(2.4)Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, S25-39 Fab (IgG1k) heavy chain, S25-39 Fab (IgG1k) light chain, ... | | Authors: | Blackler, R.J, Evans, S.V. | | Deposit date: | 2010-08-25 | | Release date: | 2011-04-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Common NH53K Mutation in the Combining Site of Antibodies Raised against Chlamydial LPS Glycoconjugates Significantly Increases Avidity.
Biochemistry, 50, 2011
|
|
4RDX
 
 | | Structure of histidinyl-tRNA synthetase in complex with tRNA(His) | | Descriptor: | ADENOSINE MONOPHOSPHATE, HISTIDINE, Histidine--tRNA ligase, ... | | Authors: | Xie, W, Tian, Q, Wang, C. | | Deposit date: | 2014-09-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for recognition of G-1-containing tRNA by histidyl-tRNA synthetase
Nucleic Acids Res., 43, 2015
|
|
1UGN
 
 | | Crystal structure of LIR1.02, one of the alleles of LIR1 | | Descriptor: | Leukocyte immunoglobulin-like receptor 1 | | Authors: | Shiroishi, M, Rasubala, L, Kuroki, K, Amano, K, Tsuchiya, N, Tokunaga, K, Kohda, D, Maenaka, K. | | Deposit date: | 2003-06-17 | | Release date: | 2004-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Extensive polymorphisms of LILRB1 (ILT2, LIR1) and their association with HLA-DRB1 shared epitope negative rheumatoid arthritis.
Hum.Mol.Genet., 14, 2005
|
|
3TTK
 
 | | Crystal structure of apo-SpuD | | Descriptor: | Polyamine transport protein | | Authors: | Wu, D.H, Lim, S.C, Song, H.W. | | Deposit date: | 2011-09-14 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural Basis of Substrate Binding Specificity Revealed by the Crystal Structures of Polyamine Receptors SpuD and SpuE from Pseudomonas aeruginosa
J.Mol.Biol., 416, 2012
|
|
4RR6
 
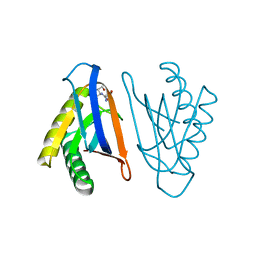 | | N-terminal editing domain of threonyl-tRNA synthetase from Aeropyrum pernix with L-Ser3AA (snapshot 1) | | Descriptor: | Probable threonine--tRNA ligase 2, SERINE-3'-AMINOADENOSINE | | Authors: | Ahmad, S, Muthukumar, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
4RRQ
 
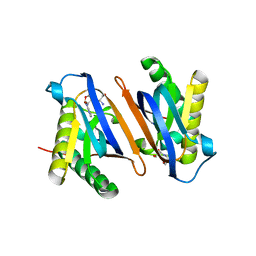 | |
2OAX
 
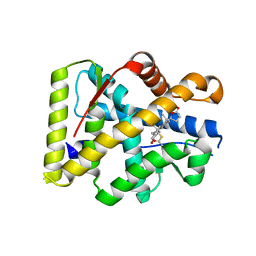 | | Crystal structure of the S810L mutant mineralocorticoid receptor associated with SC9420 | | Descriptor: | Mineralocorticoid receptor, SPIRONOLACTONE | | Authors: | Huyet, J, Pinon, G.M, Fay, M.R, Rafestin-Oblin, M.E, Fagart, J. | | Deposit date: | 2006-12-18 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural basis of spirolactone recognition by the mineralocorticoid receptor
Mol.Pharmacol., 72, 2007
|
|
1L1F
 
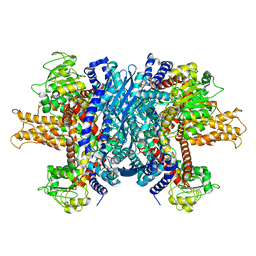 | | Structure of human glutamate dehydrogenase-apo form | | Descriptor: | Glutamate Dehydrogenase 1 | | Authors: | Smith, T.J, Schmidt, T, Fang, J, Wu, J, Siuzdak, G, Stanley, C.A. | | Deposit date: | 2002-02-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of apo human glutamate dehydrogenase details subunit communication and allostery.
J.Mol.Biol., 318, 2002
|
|
1DRE
 
 | |
2DVP
 
 | |
5EXL
 
 | |
6UJB
 
 | | Integrin alpha-v beta-8 in complex with the Fabs C6D4 and 11D12v2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C6D4 heavy chain Fab, ... | | Authors: | Campbell, M.G, Cormier, A, Cheng, Y, Nishimura, S.L. | | Deposit date: | 2019-10-02 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Cryo-EM Reveals Integrin-Mediated TGF-beta Activation without Release from Latent TGF-beta.
Cell, 180, 2020
|
|
6CD9
 
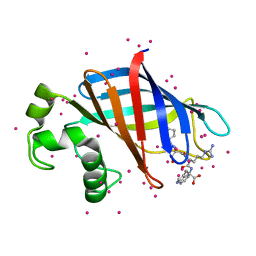 | | GID4 in complex with a peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PSRW, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
3R8D
 
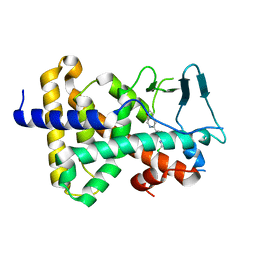 | |
1EN7
 
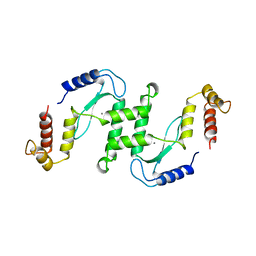 | | ENDONUCLEASE VII (ENDOVII) FROM PHAGE T4 | | Descriptor: | CALCIUM ION, RECOMBINATION ENDONUCLEASE VII, ZINC ION | | Authors: | Raaijmakers, H, Vix, O, Toro, I, Suck, D. | | Deposit date: | 1999-02-07 | | Release date: | 2000-02-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of T4 endonuclease VII: a DNA junction resolvase with a novel fold and unusual domain-swapped dimer architecture.
EMBO J., 18, 1999
|
|
5EXM
 
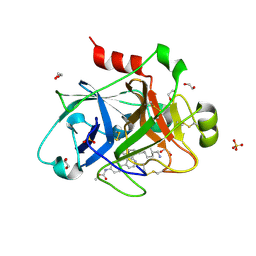 | |
2KXD
 
 | | The structure of SH3-F2 | | Descriptor: | 11-mer peptide,Spectrin alpha chain, non-erythrocytic 1,Spectrin alpha chain, non-erythrocytic 1 | | Authors: | Kutyshenko, V.P, Gushchina, L.V, Khristoforov, V.S, Prokhorov, D.A, Timchenko, M.A, Kudrevatykh, I.u.A, Fedyukina, D.V, Filimonov, V.V. | | Deposit date: | 2010-04-30 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of the chimeric protein SH3-F2
Mol.Biol.(Engl.Transl.), 44, 2010
|
|
