1UVT
 
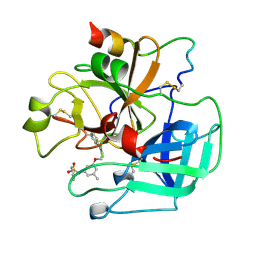 | | BOVINE THROMBIN--BM14.1248 COMPLEX | | Descriptor: | N-{3-METHYL-5-[2-(PYRIDIN-4-YLAMINO)-ETHOXY]-PHENYL}-BENZENESULFONAMIDE, THROMBIN | | Authors: | Engh, R.A, Huber, R. | | Deposit date: | 1996-10-16 | | Release date: | 1997-11-19 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzyme flexibility, solvent and 'weak' interactions characterize thrombin-ligand interactions: implications for drug design.
Structure, 4, 1996
|
|
1UXB
 
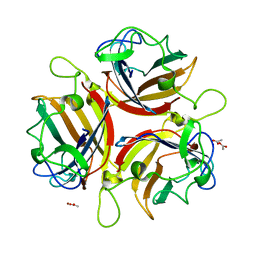 | | ADENOVIRUS AD19p FIBRE HEAD in complex with sialyl-lactose | | Descriptor: | ACETATE ION, FIBER PROTEIN, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Burmeister, W.P, Guilligay, D, Cusack, S, Wadell, G, Arnberg, N. | | Deposit date: | 2004-02-24 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Species D Adenovirus Fiber Knobs and Their Sialic Acid Binding Sites
J.Virol., 78, 2004
|
|
1UYS
 
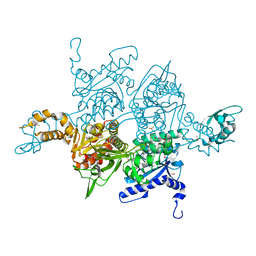 | |
1V3C
 
 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III: complex with NEU5AC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-30 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
1UZP
 
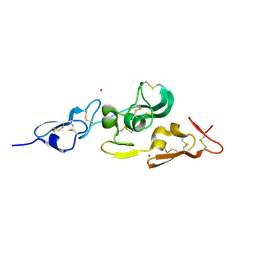 | | Integrin binding cbEGF22-TB4-cbEGF33 fragment of human fibrillin-1, Sm bound form cbEGF23 domain only. | | Descriptor: | FIBRILLIN-1, SAMARIUM (III) ION | | Authors: | Lee, S.S.J, Knott, V, Harlos, K, Handford, P.A, Stuart, D.I. | | Deposit date: | 2004-03-15 | | Release date: | 2004-04-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Integrin Binding Fragment from Fibrillin-1 Gives New Insights Into Microfibril Organization
Structure, 12, 2004
|
|
1V3R
 
 | | Crystal structure of TT1020 from Thermus thermophilus HB8 | | Descriptor: | Nitrogen regulatory protein P-II | | Authors: | Wang, H, Sakai, H, Takemoto-Hori, C, Kaminishi, T, Yamaguchi, H, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-05 | | Release date: | 2004-11-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8.
J.Struct.Biol., 149, 2005
|
|
1UXY
 
 | | MURB MUTANT WITH SER 229 REPLACED BY ALA, COMPLEX WITH ENOLPYRUVYL-UDP-N-ACETYLGLUCOSAMINE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Benson, T.E, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1996-11-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the S229A mutant and wild-type MurB in the presence of the substrate enolpyruvyl-UDP-N-acetylglucosamine at 1.8-A resolution.
Biochemistry, 36, 1997
|
|
1UWB
 
 | | TYR 181 CYS HIV-1 RT/8-CL TIBO | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 1996-11-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
1URT
 
 | | MURINE CARBONIC ANHYDRASE V | | Descriptor: | CARBONIC ANHYDRASE V, ZINC ION | | Authors: | Boriack-Sjodin, P.A, Christianson, D.W. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of an intramolecular proton transfer site in murine carbonic anhydrase V.
Biochemistry, 35, 1996
|
|
1V6T
 
 | |
1V72
 
 | |
1V8M
 
 | | Crystal structure analysis of ADP-ribose pyrophosphatase complexed with ADP-ribose and Gd | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase, GADOLINIUM ATOM | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-12 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8S
 
 | | Crystal structure analusis of the ADP-ribose pyrophosphatase complexed with AMP and Mg | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-ribose pyrophosphatase, MAGNESIUM ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-14 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8U
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E82Q mutant with SO4 and Mg | | Descriptor: | ADP-ribose pyrophosphatase, MAGNESIUM ION, SULFATE ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V9M
 
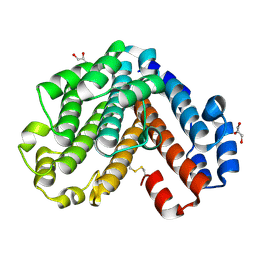 | | Crystal structure of the C subunit of V-type ATPase from Thermus thermophilus | | Descriptor: | GLYCEROL, V-type ATP synthase subunit C | | Authors: | Numoto, N, Kita, A, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the C subunit of V-type ATPase from Thermus thermophilus at 1.85 A resolution.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VCG
 
 | | Crystal Structure of IPP isomerase at P43212 | | Descriptor: | FLAVIN MONONUCLEOTIDE, isopentenyl-diphosphate delta-isomerase | | Authors: | Wada, T, Park, S.-Y, Tame, R.H, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-08 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal Structure of IPP isomerase at P43212
To be Published
|
|
1VBH
 
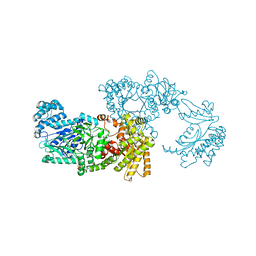 | | Pyruvate Phosphate Dikinase with bound Mg-PEP from Maize | | Descriptor: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE, SULFATE ION, ... | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
1US5
 
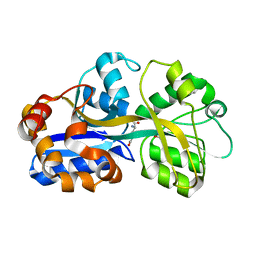 | | PUTATIVE GLUR0 LIGAND BINDING CORE WITH L-GLUTAMATE | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, PUTATIVE GLUR0 LIGAND BINDING CORE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2003-11-18 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Thermus Thermophilus Putative Periplasmic Glutamate/Glutamine-Binding Protein
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VCA
 
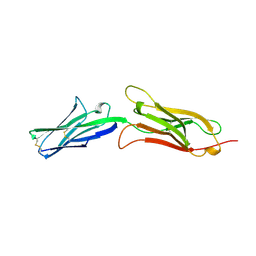 | | CRYSTAL STRUCTURE OF AN INTEGRIN-BINDING FRAGMENT OF VASCULAR CELL ADHESION MOLECULE-1 AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | HUMAN VASCULAR CELL ADHESION MOLECULE-1 | | Authors: | Jones, E.Y, Harlos, K, Bottomley, M.J, Robinson, R.C, Driscoll, P.C, Edwards, R.M, Clements, J.M, Dudgeon, T.J, Stuart, D.I. | | Deposit date: | 1995-03-21 | | Release date: | 1995-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an integrin-binding fragment of vascular cell adhesion molecule-1 at 1.8 A resolution.
Nature, 373, 1995
|
|
1UMB
 
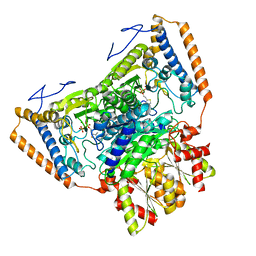 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 in holo-form | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, MAGNESIUM ION, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1USU
 
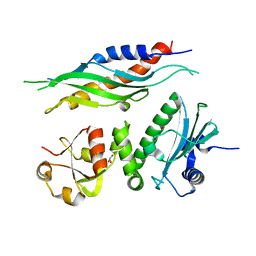 | |
1USX
 
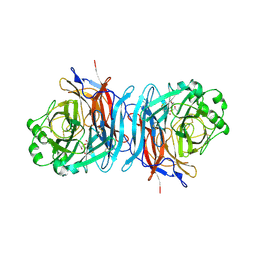 | | Crystal structure of the Newcastle disease virus hemagglutinin-neuraminidase complexed with thiosialoside | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, HEMAGGLUTININ-NEURAMINIDASE GLYCOPROTEIN, N-acetyl-alpha-neuraminic acid-(2-6)-methyl 6-thio-beta-D-galactopyranoside | | Authors: | Zaitsev, V, Itzstein, M, Groves, D, Kiefel, M, Takimoto, T, Portner, A, Taylor, G. | | Deposit date: | 2003-12-01 | | Release date: | 2004-03-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Second Sialic Acid Binding Site in Newcastle Disease Virus Hemagglutinin-Neuraminidase: Implications for Fusion
J.Virol., 78, 2004
|
|
1UNP
 
 | | Crystal structure of the pleckstrin homology domain of PKB alpha | | Descriptor: | RAC-ALPHA SERINE/THREONINE KINASE | | Authors: | Milburn, C.C, Deak, M, Kelly, S.M, Price, N.C, Alessi, D.R, van Aalten, D.M.F. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding of phosphatidylinositol 3,4,5-trisphosphate to the pleckstrin homology domain of protein kinase B induces a conformational change.
Biochem. J., 375, 2003
|
|
1UUS
 
 | | Structure of an activated Dictyostelium STAT in its DNA-unbound form | | Descriptor: | STAT PROTEIN | | Authors: | Soler-Lopez, M, Petosa, C, Fukuzawa, M, Ravelli, R, Williams, J.G, Muller, C.W. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-26 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an Activated Dictyostelium Stat in its DNA-Unbound Form
Mol.Cell, 13, 2004
|
|
1UZY
 
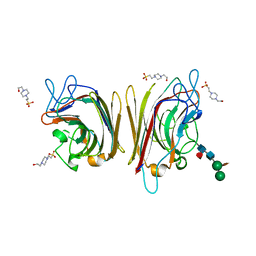 | | Erythrina crystagalli lectin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Lectin, ... | | Authors: | Turton, K, Natesh, R, Thiyagarajan, N, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2004-03-19 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Erythrina cristagalli lectin with bound N-linked oligosaccharide and lactose.
Glycobiology, 14, 2004
|
|
