3NHZ
 
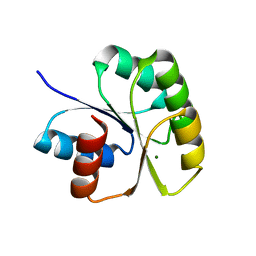 | | Structure of N-terminal Domain of MtrA | | Descriptor: | MAGNESIUM ION, Two component system transcriptional regulator mtrA | | Authors: | Barbieri, C.M, Mack, T.R, Robinson, V.L, Miller, M.T, Stock, A.M. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
3NNN
 
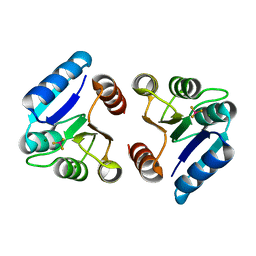 | | BeF3 Activated DrrD Receiver Domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA BINDING RESPONSE REGULATOR D, MAGNESIUM ION | | Authors: | Robinson, V.L, Stock, A.M. | | Deposit date: | 2010-06-23 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
3C97
 
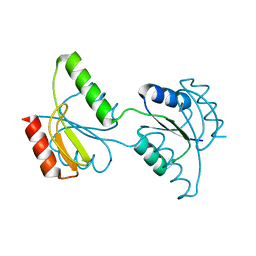 | | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae | | Descriptor: | Signal transduction histidine kinase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Chang, S, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-15 | | Release date: | 2008-03-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the response regulator receiver domain of a signal transduction histidine kinase from Aspergillus oryzae.
To be Published
|
|
3CU5
 
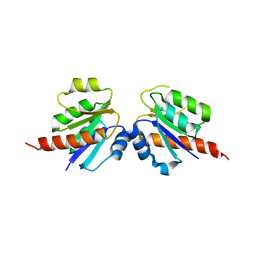 | | Crystal structure of a two component transcriptional regulator AraC from Clostridium phytofermentans ISDg | | Descriptor: | Two component transcriptional regulator, AraC family | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-15 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a two component transcriptional regulator AraC from Clostridium phytofermentans ISDg.
To be Published
|
|
3CG0
 
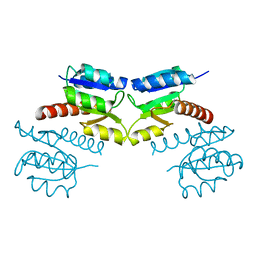 | | Crystal structure of signal receiver domain of modulated diguanylate cyclase from Desulfovibrio desulfuricans G20, an example of alternate folding | | Descriptor: | Response regulator receiver modulated diguanylate cyclase with PAS/PAC sensor | | Authors: | Patskovsky, Y, Bonanno, J.B, Romero, R, Gilmore, M, Chang, S, Groshong, C, Koss, J, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Modulated Diguanylate Cyclase from Desulfovibrio desulfuricans.
To be Published
|
|
3CFY
 
 | | Crystal structure of signal receiver domain of putative Luxo repressor protein from Vibrio parahaemolyticus | | Descriptor: | Putative LuxO repressor protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Fong, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Signal Receiver Domain of Putative Luxo Repressor Protein from Vibrio Parahaemolyticus.
To be Published
|
|
3CG4
 
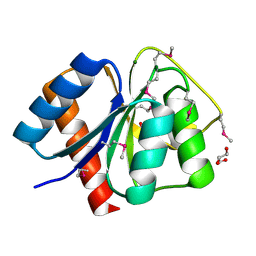 | | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Response regulator receiver domain protein (CheY-like) | | Authors: | Patskovsky, Y, Freeman, J, Hu, S, Bain, K, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-04 | | Release date: | 2008-03-11 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Response Regulator Receiver Domain Protein (Chey-Like) from Methanospirillum hungatei JF-1.
To be Published
|
|
3CHY
 
 | |
3CNB
 
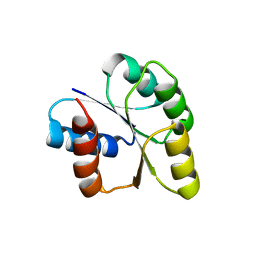 | | Crystal structure of signal receiver domain of DNA binding response regulator protein (merR) from Colwellia psychrerythraea 34H | | Descriptor: | DNA-binding response regulator, merR family | | Authors: | Patskovsky, Y, Romero, R, Freeman, J, Hu, S, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of signal receiver domain of DNA binding response regulator (merR) from Colwellia psychrerythraea 34H.
To be Published
|
|
3CZ5
 
 | | Crystal structure of two-component response regulator, LuxR family, from Aurantimonas sp. SI85-9A1 | | Descriptor: | PHOSPHATE ION, Two-component response regulator, LuxR family | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S.R, Meyer, A, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-28 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of two-component response regulator, LuxR family, from Aurantimonas sp. SI85-9A1.
To be Published
|
|
3CRN
 
 | | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1 | | Descriptor: | GLYCEROL, Response regulator receiver domain protein, CheY-like, ... | | Authors: | Hiner, R.L, Toro, R, Patskovsky, Y, Freeman, J, Chang, S, Smith, D, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-07 | | Release date: | 2008-04-22 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of response regulator receiver domain protein (CheY-like) from Methanospirillum hungatei JF-1.
To be Published
|
|
3DGF
 
 | |
3B2N
 
 | | Crystal structure of DNA-binding response regulator, LuxR family, from Staphylococcus aureus | | Descriptor: | SODIUM ION, Uncharacterized protein Q99UF4 | | Authors: | Malashkevich, V.N, Toro, R, Meyer, A.J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-10-18 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of DNA-binding response regulator, LuxR family, from Staphylococcus aureus.
To be Published
|
|
3C3M
 
 | | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Dickey, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1.
To be Published
|
|
3A0U
 
 | | Crystal structure of response regulator protein TrrA (TM1360) from Thermotoga maritima in complex with Mg(2+)-BeF (wild type) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator, ... | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A10
 
 | | Crystal structure of response regulator protein TrrA (TM1360) from Thermotoga maritima in complex with Mg(2+)-BeF (SeMet, L89M) | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Response regulator, ... | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-25 | | Release date: | 2009-10-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3NNS
 
 | | BeF3 Activated DrrB Receiver Domain | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA BINDING RESPONSE REGULATOR B, MAGNESIUM ION | | Authors: | Robinson, V.L, Stock, A.M. | | Deposit date: | 2010-06-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of response regulator autophosphorylation through interdomain contacts.
J.Biol.Chem., 285, 2010
|
|
3OO1
 
 | |
3OO0
 
 | | Structure of apo CheY A113P | | Descriptor: | AMMONIUM ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Guanga, G.P, Miller, P.J, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2010-08-30 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Exploring the effect of an allosteric site on conformational coupling in CheY
To be Published
|
|
3OLV
 
 | |
3OLY
 
 | |
3OLW
 
 | |
3OLX
 
 | |
3RVK
 
 | | Structure of the CheY-Mn2+ Complex with substitutions at 59 and 89: N59D E89Q | | Descriptor: | Chemotaxis protein CheY, MANGANESE (II) ION | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVP
 
 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89K | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
